Abstract
Current screening procedures for colorectal cancer are imperfect, highly invasive and result in increased mortality rates due to low compliance. The goal of the experiments reported herein is to identify potential blood-based biomarkers indicative of early-stage intestinal cancers using the ApcMin/+ mouse model of intestinal cancer as an experimental system. Serum proteins from tumor-bearing ApcMin/+ mice were quantitatively compared to tumor-free Apc+/+ wild-type mice via in anima metabolic labeling with 14N/15N labeled Spirulina algae and an LTQ Orbitrap mass spectrometer. Out of 1116 total serum proteins quantified, this study identified 40 that were differentially expressed and correlated with the increase in intestinal neoplasms. A subset of these differentially expressed proteins underwent a secondary quantitative screen using selected reaction monitoring-mass spectrometry with stable isotope-labeled peptides. Using both quantitative techniques, we identified MGAM and COL1A1 as downregulated and ITIH3 and F5 as upregulated in serum. All but COL1A1 were similarly differentially expressed in the mRNA of neoplastic colonic tissues of ApcMin/+ mice compared to normal wild-type tissue. These differentially expressed proteins identified in the ApcMin/+ mouse model have provided a set of candidate biomarkers for future validation screens in humans.
Keywords: Blood protein biomarkers, Intestinal cancer, Colon cancer, ApcMin/+ mouse, 14N/15N metabolic labeling, Tandem mass spectrometry, Selected Reaction Monitoring-Mass Spectrometry, mRNA Microarray
Introduction
Colorectal cancer is the second most common cause of cancer-related death in the United States and in many other Westernized nations.1, 2 While early detection significantly increases the chance of survival, compliance with recommended screening guidelines is low, resulting in late-stage diagnosis and consequent high mortality rates.3 The American Cancer Society and American Gastroenterology Society have identified colonoscopy as the most sensitive and specific screen and recommend the procedure once every ten years over the age of fifty.4–6 For patients who are not candidates for colonoscopy, these agencies recommend other procedures such as sigmoidoscopy or Computed Tomographic Colonography (CTC) scanning every five years. Due to the lack of compliance with these and other recommended invasive detection screens, clinicians and researchers are striving to identify diagnostic procedures for early colon cancer detection that do not require bowel preparation and are less invasive.7, 8
Roughly 85% of human colon cancers are initiated by mutations in the gatekeeper tumor suppressor gene Adenomatous Polyposis Coli (APC).9–11 APC encodes a very large protein of more than 2,800 amino acids that acts as a tumor suppressor in the well-studied WNT cell-signaling pathway. In an inactive WNT pathway, cytoplasmic APC binds to the transcription factor β-catenin, sequesters it in the cytoplasm, and targets the protein for degradation by the 26S proteasome. Mutations in APC can result in decreased degradation of β-catenin and its increased transport to the nucleus. This increased transport results in constitutive transcriptional activation of genes involved in cell growth and proliferation, which are inherent to the formation of colonic neoplasms.9
The Multiple intestinal neoplasia (Min) mouse model of familial intestinal cancer develops multiple intestinal and colonic neoplasms, and has been commonly used to study mutations in APC over the last two decades.12 The loss of APC function in these Min mice is caused by a nonsense mutation at amino acid residue 850 that causes premature termination of translation of the mRNA and a significantly truncated protein. In this study, we have applied in anima 15N labeling with tandem mass spectrometry to quantify early cancer-induced changes in the serum proteome of this well-studied model for intestinal neoplasia.
We have previously shown the success of in anima metabolic labeling using the 15N stable nitrogen isotope as a quantitative proteomic approach to identify differentially expressed proteins in colonic tumor tissue from Min mice compared to wild type Apc+/+ mice.13 In this experiment, mice were fed diets in which the only source of nitrogen was derived from Spirulina algae grown under conditions to incorporate the heavy isotope 15N at either natural abundance (0.364%) or enriched to over 98% 15N within all nitrogen-containing cellular components. In anima metabolic labeling allows differentially labeled biological material to be combined in equal parts immediately upon sample collection to provide a sophisticated internal control during multi-step sample preparations.14
In recent years ‘omics-based technologies have opened new avenues to study the molecular basis of cancer and have potential applications in cancer drug development and detection.15, 16 The motivation behind much of the detection research is the desire to develop a minimally invasive screen that is both sensitive and specific for a disease indication. Blood-based screening is preferred owing to ease of sample collection and consequently increased potential for patient compliance. However, in the case of blood-based biomarker discovery studies, a major challenge lies in overcoming the ten orders of magnitude in dynamic range of all of the various blood proteins.15 Multiple separations with affinity chromatography followed by another form of separation prior to reversed-phase LC-MS/MS are typically used to address the dynamic range problem.17, 18 When samples are prepared separately, these multi-step sample preparations have the drawback of variable sample handling and loss, thus introducing greater error into the quantitation of differentially expressed protein levels. In the in anima labeling procedure used herein, combining equal volumes of blood from one wild type and one mutant Min mouse immediately upon collection, provides an inherent internal control for error in this process and significantly reduces false positive biomarker identifications.
To further assess the validity of putative biomarker identifications, a secondary protein quantification screen is necessary. We have chosen to use selected reaction monitoring-mass spectrometry (SRM-MS) as a method to further verify differential expression of a subset of our biomarker candidates.19, 20 SRM is a superior quantification technique compared to the Western blotting and ELISA assays often used in secondary quantification screens in that it does not rely on antibodies that may cross-react with other proteins in the sample. Additionally, SRM allows for multiplexing of many peptides corresponding to many protein candidates in a single assay, thus increasing the number of proteins monitored.
Here we present a study that uses in anima metabolic labeling of Min and wild-type mice to assess changes of protein expression in serum. This study applies metabolic labeling as an internal control, and analyzes serum from the same mice at three time points to observe changes in protein levels over time. Two other biological sets of Min and WT mice were generated for the purpose of mRNA microarrays of tumor tissue compared to normal epithelium and SRM-MS analysis of selected serum protein candidates. The goal of this research is to develop a set of differentially expressed blood protein candidates for future biomarker screening of early-stage intestinal cancers.
Experimental Procedures
Mouse metabolic labeling study
Metabolic labeling of mice
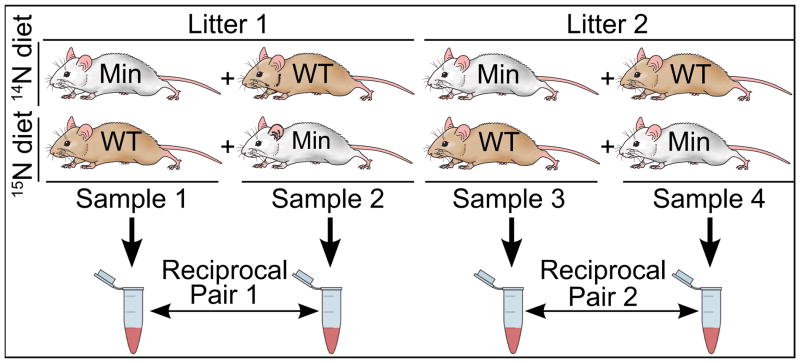
A detailed description of the metabolic labeling for these mice can be found in the supplementary materials of our previously published work.13 Briefly, four female wild type mice and four female Min mice from two litters were grouped to create four sets of samples, each containing one wild-type mouse and one Min mouse from the same litter, as shown in Figure 1. Starting at 22 days of age, weaned mice were fed a diet of either 99.636%/0.364% 14N/15N (natural abundance) or >98% 15N-enriched Spirulina. Both diets were supplemented with essential, non-nitrogenous fats and nutrients to ensure animal health and palatability of the food source. For Samples 1 and 3, the Min mouse was fed the natural abundance 14N natural abundance diet and the wild type littermate was fed the 15N Spirulina diet. A reversed metabolic label was applied to Samples 2 and 4 to create a reciprocal metabolic labeling of littermates to control for potential diet-related protein expression changes.
Figure 1.
Mouse metabolic labeling schema. Eight mice were fed diets of >98% 15N or natural abundance 14N Spirulina starting at 20 days of age. Blood was collected at 40, 52, and 66 days of age from each mouse and combined in equal amounts to form four 14N/15N samples. Reciprocal metabolic labels were used in each litter to make two reciprocally labeled samples that controlled for isotopic effects.
Blood sample collection
Whole blood (0.4 ml) was collected from each mouse at each time point using a retro-orbital sinus blood collection technique with anesthesia, according to a protocol approved by the Animal Care and Use Committee of the University of Wisconsin School of Medicine and Public Health. Whole blood was collected at three time points for each mouse: 40 days of age (18 days post-weaning), 52 days of age (29–33 days post-weaning) and 66 days of age (43–46 days post-weaning). Equal volumes of blood from one Min mouse and one wild-type mouse (one fed a Spirulina diet of natural abundance nitrogen and the other a >98% 15N diet) were immediately combined upon collection, prior to clotting and centrifugation.
Serum protein isolation and depletion
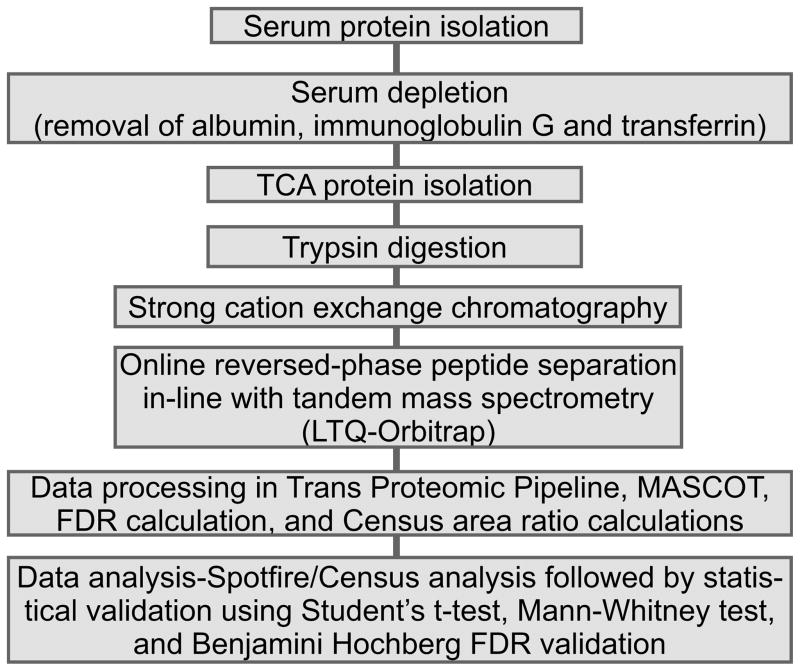
Figure 2 summarizes the sample preparation and data analysis procedures, as described in detail below. Fresh 14N/15N blood samples were clotted on ice for 30 minutes and then centrifuged at 2,000×g for 20 minutes at 4°C. Three layers formed consisting of lipid (top), serum protein (middle), and red blood cells (pellet). The protein layer was reserved and the others discarded. The protein fraction was washed five times with 30 kDa MWCO Amicon Centriprep units with 5 ml of 20% acetonitrile/80% MilliQ H2O at 1500 g for 1 hour at 4°C. The serum samples were frozen, lyophilized, and stored at −80°C.
Figure 2.
Flow of experimental methods for the 14N/15N serum samples. After combining the blood samples in equal amounts and forming serum, all mouse samples were prepared and analyzed using the general methods described.
The major blood proteins (albumin, transferrin, and IgG) were removed using a 4.6×100 mm Agilent Multiple Affinity Removal Column using all of the proprietary buffers and filters described in the manufacturer’s instructions. A Waters 1740 HPLC equipped with a manual sample injector was used. Prior to depletion, the lyophilized serum sample was solubilized in Agilent buffer injected onto the HPLC. Each injection was monitored at 215 and 280 nm using a Waters 2996 photodiode array detector with both bound and unbound fractions collected. Fractions were stored at −80°C until further use.
Protein precipitation and trypsin digestion
Unbound samples from serum depletions were thawed on ice and concentrated in Agilent 5 kDa MWCO concentrating filter units at 4,000×g, at 4°C until ≤500 μl liquid remained. Concentrated protein was immediately precipitated by adding cold trichloroacetic acid to a final concentration of 10% and allowing the samples to incubate on ice for five minutes. Proteins were pelleted using a microcentrifuge for five minutes at 4000×g. The centrifugation step was repeated with five washes of cold 80% acetone in water followed by one wash in cold 50% methanol in water. The supernatant was removed after each wash and the pellet allowed to air dry after the final wash. Pellets were resolubilized overnight at 4°C in 40 mM ammonium bicarbonate, 8 M urea, pH 8. The partially dissolved pellets were sonicated for five-second pulses a total of five times using a sonicating probe followed by dilution of the highly concentrated urea to 1.6 M using 40 mM ammonium bicarbonate, pH 8.
Prior to trypsin digestion, a BCA protein concentration assay (Pierce) was performed according to the manufacturer’s instructions. Up to 600 μg serum protein from each sample was digested using sequencing grade porcine trypsin (Promega) at a 1:50 trypsin-protein ratio in the presence of 5 mM DTT. Each digestion was performed at 37°C overnight followed by acidification to pH 2–3 using TFA to stop the proteolytic reaction. Waters 1 cc tC18 (100 mg sorbent) Sep-Pak® columns were used to desalt the digested peptides according to manufacturer’s instructions. Following sample elution, the peptides were dried using a vacuum centrifuge and stored at −80°C until use.
Strong cation exchange chromatography
Dried peptides were resolubilized in 500 μl of 10 mM KH2PO4, pH 3.0 and 20% acetonitrile (buffer A) and separated by strong cation exchange chromatography (SCX) on a Waters 1740 HPLC equipped with a Gilson model 201c fraction collector and a PolyLC A2555C column. Following a five-minute wash with buffer A, a 45 minute linear gradient reaching a final concentration of 100% buffer B (0.4 M NaCl, 10 mM KH2PO4, pH 3.0 and 20% acetonitrile) was applied. Remaining peptides were washed off of the column for ten minutes at 100% buffer B followed by an immediate switch to a ten minute column reconditioning with buffer A. One fraction was collected each minute for a total of 60 fractions at a flow rate of 1 ml/min. For each of the biological replicates at each time point, 60 SCX fractions were combined into a final 6–18 fractions for mass spectrometry analysis depending on the amount of starting material. The fractions were combined based on both the complexity of the UV traces at 215 nm and 280 nm and based on a rudimentary analysis of fraction complexity using an MDS SciEX 4800 MALDI TOF/TOF. In total, 180 strong cation exchange fractions were used across all samples and time points for LC-MS/MS analysis. SCX fractions were flash frozen in liquid nitrogen, lyophilized, and desalted using Spec PT C18 solid-phase extraction pipette tips (Varian) according to the manufacturer’s protocol. Following sample elution, the peptides were dried using a vacuum centrifuge and stored at −80°C until use.
Mass spectrometry data collection
Peptides were resolubilized in 0.1% formic acid in water (reversed-phase buffer A) and an estimated 8 μg loaded onto a 15–20 cm home-packed Magic C18 column for a 240-minute reversed-phase gradient separation in-line with an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific). The flow rate was set at 0.2 μl/min for the entire analysis. The first 20 minutes were run at 1% Buffer B (95% acetonitrile, 0.1% formic acid, 4.9% water) followed by a linear increase in the gradient to 40% buffer B at 215 minutes. A steeper gradient was then applied to reach 60% buffer B at 235 minutes and 100% B at 240 minutes. The column was washed with buffer B for three minutes followed by an immediate gradient change to 1% buffer B for a fifteen minute column reconditioning. A data-dependent acquisition method was used, selecting the top five most intense precursor masses observed in the Orbitrap with MS/MS spectra obtained in the linear ion trap. A dynamic exclusion of precursor masses was applied for 40 seconds after one repeat MS/MS analysis. A “blank” 45-minute gradient was run between each SCX run and sample injections alternated between 15N-Min/14N-WT SCX fractions and matching reciprocal partnered SCX fractions. There were no technical replicates for Orbitrap MS analysis.
Thermo “.raw” files were converted in the Trans Proteomic Pipeline (TPP) to a suitable format (.raw to .mzXML to .mgf) for database searching using the MASCOT search algorithm.21 All files were searched against the Mus musculus NCBI database (version as of 3/14/2011, 30,062 protein sequences) allowing for up to two missed cleavages, and considering oxidized methionines and deamidation of aspartate and glutamate as variable modifications. Search parameters were for tryptic peptides, a precursor mass tolerance of +/−2.5 Da and a product ion mass tolerance of +/−0.5 Da. The database was modified to include sequences of common contaminants such as trypsin and human keratins. All initial searches were done in MASCOT using the natural mass of nitrogen. For some samples, a second 15N search was later done in which the added mass of the neutron was incorporated. This was done for SCX fractions in which proteins that were identified with very high 14N ratios in one mouse sample may not have been identified in a database search in the reciprocally labeled sample due to a low relative 14N peak. MASCOT output files were converted to “.pepXML” using TPP and were filtered to a 1% false discovery rate at the peptide level using a ‘decoy’ reversed-protein sequence database and in-house software.
The filtered 14N MASCOT search results were processed to identify the average percent 15N enrichment of the blood proteins, using the Census software package.22 This was done to identify the isotopologues that combine to form the clusters of isotopic peaks upon which our quantitative calculations are based. Such a calculation is necessary given that 100% 15N incorporation could not be achieved in blood over the time in which samples were collected. Using the average incorporation ratio, the data were reprocessed in Census to obtain the quantitative ratio measurements by extracting ion chromatograms within a 30-ppm mass window around the 14N and 15N isotopic envelopes. For each mouse sample at each time point, a histogram of the Census area ratios was generated. A correction factor was applied to center the results at a 1:1 ratio.
Data analysis
Peptide area ratios from all mouse samples and time points were collated into Spotfire (TIBCO) software where box plots were generated to assess general quantitative changes and variance trending across the samples and time points for each quantified protein (Supplementary Figure 1). Each box plot displayed all of the area ratios (Min/wild type) for all of the peptides of a single protein for a single mouse sample at a single time point. The box plots for a single protein were displayed together for all samples and time points to determine the trending changes over time and across the 14N/15N mouse serum samples. Box plots were generated using the default settings in Spotfire, and proteins that appeared to show the same change in protein expression between reciprocal members of a pair were marked for manual analysis in Census.
Seemingly anomalous peptide quantifications were subjected to manual validation of Census extracted ion chromatograms and data points were excluded when they were judged to be unreliable. Peptides were rated as “good”, “fair” or “poor” with only poor peptides being removed from the data set. A peptide was deemed “poor” if it displayed an incorrect elution time, poor signal intensity resulting in a poor chromatogram, or an assigned peptide sequence whose proteotypic properties were deemed incompatible with those of numerous other peptides for that protein. Those peptides passing manual inspection were then grouped by protein. The geometric mean of the log2 area ratios and the standard error were calculated for each mouse sample and timepoint. Statistical validation was done using an in-house R-script that used the non-parametric Mann-Whitney test to obtain U-values. The U-values were further filtered using the Benjamini-Hochberg q-test to remove false positive values at a false discovery rate of 5% or less (q<0.05). The U-test was considered significant if the resultant U-value fell below 0.05 after Benjamini-Hochberg filtering.
The data generated from this discovery study also contained numerous low replicate candidates that were identified in only one or two samples at a single time point, and single identifications that were therefore not eligible for statistical analysis. While many of these outlying candidates likely represent errors in peptide and protein identification or quantification, at least some of these protein identifications may represent proteins that are bona fide candidate protein biomarkers. To help address this possibility, all singly identified peptides that appeared to be changing in expression level by two-fold or more (log2 ratio of 1.0 or greater) were manually inspected in Census and in the raw MS1 and MS/MS spectra to assure the quality of the identification and quantification. These peptides were also validated for uniqueness through BLAST searching.23
To represent this large data-set adequately, the putative biomarker results are presented in three categories: high replicate with high statistical confidence; high replicate with reduced statistical confidence; and low replicate with little or no statistical evaluation available. High replicate, high statistical confidence proteins were identified as being statistically differentially expressed in at least three out of four samples at the 52 and/or 66 day time points. Statistical significance was defined as having a U-value less than 0.05 and a corresponding q-value less than 0.05. The category with high replicates but reduced statistical confidence included proteins that had statistically relevant differential expression in only one reciprocally labeled sample, or single peptide identifications where no statistical calculations could be made. The proteins with low replicate results had little or no statistical substantiation. The proteins in this category either displayed no reciprocal sample validation, or the protein had only a single unique peptide hit changing at a log2 ratio of 1.0 or greater.
SRM-MS Screen
Mouse sample preparation
Blood from total of ten C57BL/6-6J female mice, 5 ApcMin/+ and 5 Apc+/+, was collected at 60 days of age as previously described. Mice were raised on 5020 breeder chow (Purina, St Louis, MO) and therefore all proteins were of natural 14N abundance. Serum protein isolation and depletion were done as previously described and trypsin digestion was done as described but in the presence of a fixed amount of stable isotope labeled peptide standards that were added just prior to digestion.
LC-SRM-MS method
All peptide reference standards were synthesized by the UW-Madison Biotechnology Center’s peptide synthesis core facility with the incorporation of one 13C15N labeled amino acid. Peptides were resolubilized in 0.1% formic acid, 5% acetonitrile, and water to a concentration of 1 μg/μl endogenous peptides. Liquid chromatography separation was achieved using a NanoLC ultra 2D (Eksigent) equipped with a nanoflex cHiPLC. The microfluidic chip was a 75μm diameter 15cm length column with C18 3μm resin at a 120Å pore size and the temperature of the cHiPLC system was set to 37°C. A 90 minute gradient at a flow rate of 300nl/min was applied as follows: starting conditions were set at 97% 0.1% formic acid in water (buffer A) and 3% 0.1% formic acid in acetonitrile (buffer B) and increased linearly to 15% B by 30 minutes. Buffer B was increased linearly to 35% by 60 minutes and then a steeper gradient to 50% B was applied to 85 minutes. The gradient was switched back to starting conditions at 90 minutes. Peptides were eluted directly into a 5500 QTrap (AbSciex). Peptide precursors were selected in Q1 followed by fragmentation in q2 and subsequent monitoring of the top 3–4 transitions for each peptide in Q3. All Q1 and Q3 masses were measured at unit resolution. To maximize dwell times, a 5-minute scheduling window was applied with a 1.5 second cycle time. Method development and peak analysis was done using Skyline software.24
Data analysis
All SRM results were imported into Skyline and peaks integrated. Technical replicates with poor peak shapes and intensities relative to other technical replicates were discarded. A minimum of 3 technical replicates per biological sample were reserved for evaluating each peptide. Peak areas from the top 3–4 transitions were exported from Skyline and further analyzed for interferences using AuDIT.25 The transition with the lowest coefficient of variance (%CV) for each peptide was identified and used to quantify the ratio of the endogenous peptide compared to the reference standard in Skyline. The area ratios of endogenous to internal reference standard for all technical replicates from a single biological sample were averaged. The medians were calculated for the Min biological samples and the wild type biological samples. The biological replicate whose area ratio was furthest from the median was removed, and the four remaining area ratios from the biological replicates were averaged. A ratio of Min/WT was calculated for each of the peptides and a student’s t-test was used to determine statistical significance.
mRNA isolation and analysis
Four male (C57BL/6J-ApcMin/+ x BTBR) F1 Min mice were aged to 110 days and then sacrificed. At sacrifice, the entire intestinal tract was removed, opened longitudinally and rinsed with PBS. Samples from all tumors as well as normal epithelial scrapings were taken from each animal and stored in Qiagen RLT Plus lysis buffer. RNA was isolated from similarly sized tumors and normal epithelium from the central region of the colon. The area within 4 mm of any tumor was avoided to prevent contamination with hyperplastic tissue. DNA and RNA were isolated concurrently using the Qiagen All-prep Mini-kit. Once isolated, optical density readings and concentrations of both the DNA and RNA were determined using a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific) and RNA quality was determined using a Agilent 2100 Bioanalyzer. The McArdle Laboratory for Cancer Research Microarray facility collected data for each sample on Agilent Mus musculus 8×60k (G4852A) gene expression microarrays. Data files were loaded into Partek Genomics Suite software for analysis.
Results
In anima metabolic labeling study
The time points chosen for serum in the in anima metabolic labeling study reflect time points early in the formation of neoplastic polyps in the Min mouse, as determined by tumor multiplicity rates measured in past studies.12, 26 The time points were not only early stage according to historical tumor multiplicity results, but also prior to the onset of any visible signs of anemia, which is known to occur at later stages of disease in these mice. For each mouse, the general health was visually assessed and the weight measured daily. No discernible differences were observed between Min and wild type mice or between mice fed on the 14N or 15N Spirulina diets. The percent 15N incorporation for each 15N labeled mouse was determined at each time point to ensure that the enrichment level exceeded the minimum of 70% 15N required by Census for reliable quantification. The 15N incorporation for mice on the 15N Spirulina diet was 88–90% at 40 days of age, 91–92% at 52 days, and 93–95% at 66 days of age (Supplementary Figure 2), thus enabling Census quantification at all time points.
In total, 1,116 proteins were quantified across 180 strong cation exchange fractions comprising the entire 14N/15N dataset. Table 1 summarizes the level of quantitative data obtained; presenting the number of times the proteins were identified across the four mouse samples and three time points. More than half of the proteins were identified in at least one reciprocal sample (52.43%) across multiple time points and can be attributed to the fact that many are higher abundance serum proteins. Approximately 37% of the proteins contained only one peptide from that protein at a single time point. We attribute this to the lower relative abundance of these proteins in serum. For example, in this study alpha-fetoprotein was identified with only two unique peptide hits at one time point (expression changes were not statistically significant) and is a protein known to be present at low-levels (around 3 ng/ml) in serum from human adults.27 The number of singly quantified data points is to be expected given the 1010 dynamic range of blood protein concentrations, and given that it has been well established that reproducibility in protein identifications in a complex shotgun proteomics experiment is usually between 35–60% between replicate injections.28, 29 Newer search algorithms such as MS-GF and MyriMatch may be beneficial in future database searches to improve the number of protein identifications.30, 31 In order to use as much of the collected data as possible for biomarker analysis, the data was subdivided into three categories with ranked levels of statistical confidence as described in the Methods.
Table 1.
Number of proteins quantified across all four mouse samples and time points
| Number of samples | Number of time points
|
||||
|---|---|---|---|---|---|
| 3 | 2–3 | 2 | 1–2 | 1 | |
| 4 | 138 (12.36%) | 111 (9.95%) | 14 (1.25%) | 84 (7.53%) | 8 (0.72%) |
| 3 | 0 | 10 (0.90%) | 9 (0.81%) | 85 (7.62%) | 50 (4.48%) |
| 2 (reciprocal pairs) | 0 | 0 | 5 (0.45%) | 18 (1.61%) | 63 (5.65%) |
| 2 (non-reciprocal pairs) | 0 | 0 | 1 (0.01%) | 22 (1.97%) | 67 (6.00%) |
| 1 | 0 | N/A | 14 (1.25%) | N/A | 417 (37.36%) |
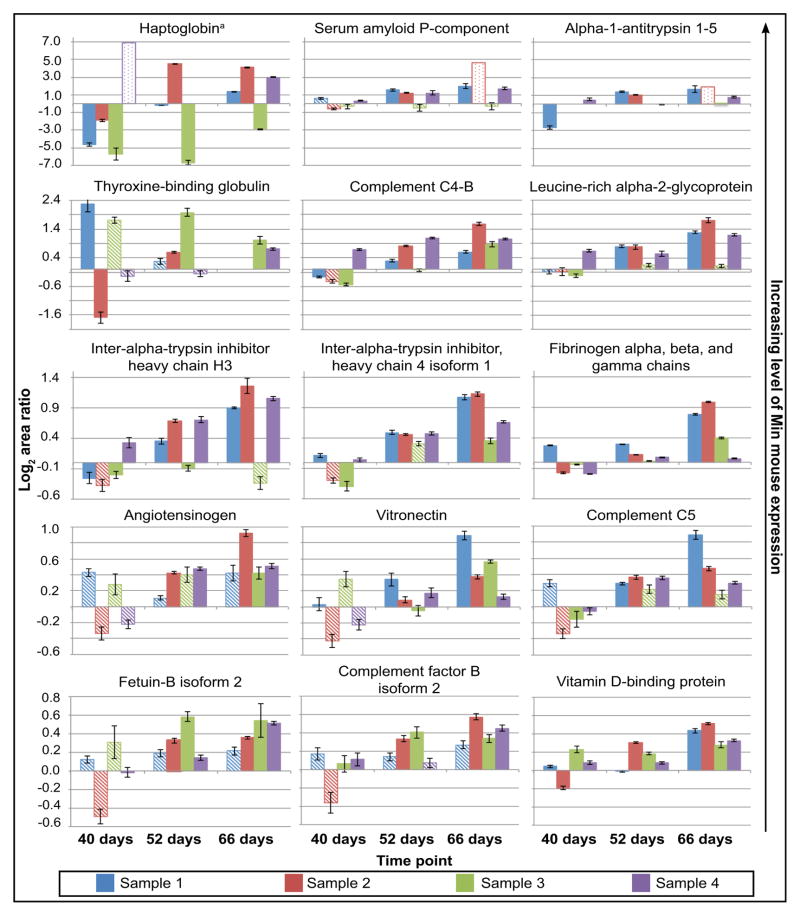
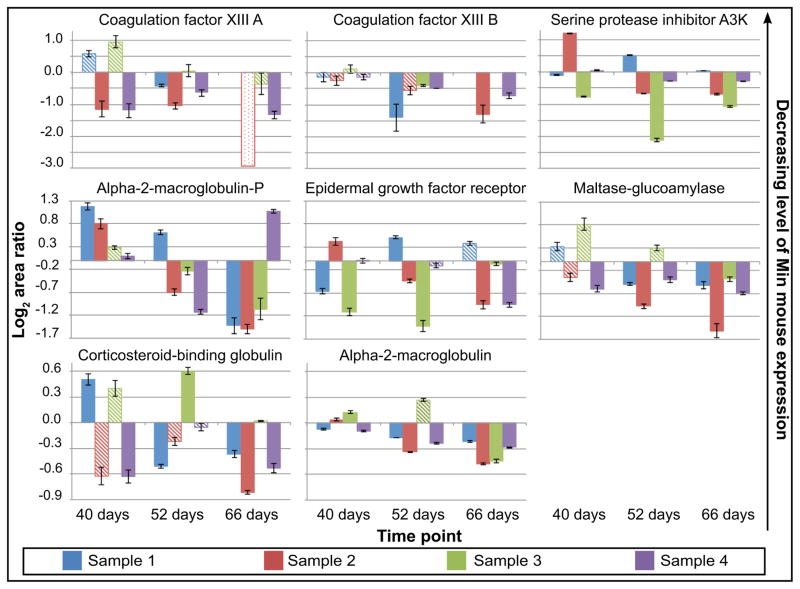
Using the 14N/15N metabolic labeling strategy, 40 (~3.6%) of the blood proteins identified in this study were differentially expressed with statistical significance at some stage in the development of intestinal neoplasms in Min mice. These proteins were identified across more than one mouse sample, and were confirmed by a reciprocally labeled mouse sample. Figure 3 and Table 2 (see also Supplementary table 1) summarize the identification of fifteen proteins that showed upregulation in Min mice compared to wild-type mice. Though mouse-to-mouse variability was observed, these proteins exhibited upregulation nearly two-fold or higher in at least three of four replicates during the 52-day and/or 66-day time point. Interestingly, eight proteins were also identified as downregulated in at least three out of four mouse samples at 52 and/or 66 days with high statistical confidence (Figure 4 and Table 2). A second set of proteins exhibited differential expression with reduced statistical confidence (Table 3, Supplementary Table 2). A total of 17 proteins were differentially expressed in this category but did not change as significantly in expression level compared to those in Figures 3 and 4. However, these proteins show statistically significant differential expression in many cases.
Figure 3.
Upregulated proteins in the 14N/15N Min mouse displaying high statistical confidence at 52 and/or 66 days in at least three out of four mouse samples. Solid bars on the bar chart represent the geometric mean of the log2 area ratios (all ratios are shown as Min/WT) that are statistically significant. Significance was defined as a U-value less than 0.05 and corresponding q-values less than 0.05. Hashed bars represent area ratios that do not pass statistical cutoffs for significance. Outlined bars with light dots represent values where there is no statistical data due to the presence of only one data point.
aMASCOT database searches for the reciprocal 15N labeled proteins show substantial increases in this protein in sample 4 at 52 days although this data is not represented in this graph (all values shown in the figure are from 14N MASCOT searches).
TABLE 2.
Proteins differentially expressed in Min compared to wildtype with high statistical confidence
| Protein name | Gene symbol | NCBI number | Log2 Area ratio Min/WTa | U-valuea | mRNA expression Min mouse (Tumor/Normal Epithelium)b | Microarray P-valueb |
|---|---|---|---|---|---|---|
| Haptoglobin | Hp | NP_059066.1 | 3.00 | 3.55E-124 | Tumors up 63.5x | 1.2091E-05 |
| Serum amyloid P-component | Apcs | NP_035448.2 | 1.55 | 9.92E-06 | Tumors down 3.5x | 1.3623E-05 |
| Alpha-1-antitrypsin 1–5 | Serpina1e or Serpina1 | NP_033273.1 | 1.41 | 1.06E-22 | Tumors down 1.5x | 0.0141238 |
| Thyroxine-binding globulin | Serpina7 | NP_808588.3 | 0.58 | 4.24E-17 | Not Significant | |
| Complement C4-B | C4b | NP_033910.2 | 0.80 | 7.38E-71 | Not Significant | |
| Leucine-rich alpha-2-glycorprotein | Lrg1 | NP_084072.1 | 1.28 | 5.74E-09 | Tumors up 110x | 7.7957E-07 |
| Inter-alpha-trypsin inhibitor heavy chain H3 | Itih3 | NP_032433 | 0.69 | 2.73E-16 | Tumors up 4.6x | 0.00128899 |
| Inter-alpha-trypsin inhibitor heavy chain 4, isoform 1 | Itih4 | NP_061216.2 | 0.67 | 1.72E-77 | Not Significant | |
| Fibrinogen alpha, beta, and gamma chains | Fga, Fgb, Fgg |
Alpha: NP_034326, NP_001104518.1 Beta: NP_862897.1, Gamma: NP_598623.1 |
0.99 | 1.15E-108 | Not Significant | |
| Angiotensinogen | Agt | NP_031454.3 | 0.42 | 2.54E-15 | Receptor (Agtr1a) down 13x in tumors | 2.4072E-06 |
| Vitronectin | Vtn | NP_035837.1 | 0.89 | 6.08E-11 | Not Significant | |
| Complement C5 | Hc or C5 | NP_034536.1 | 0.36 | 2.51E-23 | Not Significant | |
| Fetuin-B isoform 2 | Fetub | NP_001077373.1 | 0.51 | 8.78E-20 | Tumors up 18x | 2.7666E-05 |
| Complement factor B isoforms 1 and 2 | Cfb | NP_001136178.1 | 0.45 | 8.41E-38 | Not Significant | |
| Vitamin D-binding protein | Gc | NP_032122.1 | 0.30 | 9.00E-167 | Not detected | |
| Coagulation factor XIII A | F13a1 | NP_001159863, NP_083060 | −1.34 | 1.74E-10 | Not Significant | |
| Coagulation factor XIII B | F13b | NP_112441.2 | −0.40 | 6.91E-05 | Tumors down 1.2x | 0.00113888 |
| Serine protease inhibitor A3K | Serpina3k | NP_035588.2 | −0.29 | 9.37E-85 | Not Significant | |
| Alpha-2-macroglobulin-P | A2m | NP_783327.2 | −1.13 | 2.29E-23 | Not Significant | |
| Epidermal growth factor receptor, isoform 1 | Egfr | NP_997538.1 | −1.42 | 1.25E-09 | Not detected | |
| Maltase-glucoamylase | Mgam | NP_001164474.1 | −0.69 | 2.26E-20 | Tumors down 2x | 0.0056434 |
| Corticosteroid-binding globulin | Serpina6 | NP_031644.1 | −0.51 | 7.98E-14 | Not Significant | |
| Alpha-2-macroglobulin | Pzp | NP_031402.3 | −0.22 | 7.74E-219 | Not Significant |
Value represents the the area ratio at 52 or 66 days with the lowest U-value. To see all area ratios and results, see the supplementary data.
Significance was defined as a p-value of less than 0.05.
Figure 4.
Downregulated proteins in the 14N/15N Min mouse displaying high statistical confidence at 52 and/or 66 days in at least three out of four mouse samples. Solid bars on the bar chart represent the geometric mean of the log2 area ratios (all ratios are shown as Min/WT) that are statistically significant. Significance was defined as a U-value less than 0.05 and corresponding q-values less than 0.05. Hashed bars represent area ratios that do not pass statistical cutoffs for significance. Outlined bars with light dots represent values where there is no statistical data due to the presence of only one data point.
TABLE 3.
Proteins differentially expressed in Min compared to wildtype with reduced statistical confidence
| Protein name | Gene symbol | NCBI number | Log2 Area ratio Min/WTa | U-valuea | mRNA expression Min mouse (Tumor/Normal Epithelium)b | Microarray P-valueb |
|---|---|---|---|---|---|---|
| sulfhydryl oxidase 1 isoform a | Qsox1 | NP_001020116.1 | 0.64 | 2.01E-07 | Not Significant | |
| complement factor I | Cfi | NP_031712.2 | 0.61 | 2.47E-03 | Tumors up 879x | 1.28E-09 |
| C-reactive protein | Crp | NP_031794.3 | 0.26 | 5.05E-08 | Not Significant | |
| cathepsin E | Ctse | NP_031825.2 | 2.81 | 3.12E-03 | Not Significant | |
| coagulation factor V | F5 | NP_032002.1 | 0.32 | 1.33E-07 | Tumors up 6.3x | 5.49E-04 |
| serine protease inhibitor A3N | Serpina3n or Serpina3 | NP_033278.2 | 0.38 | 4.68E-03 | Tumors up 3.6x | 3.54E-03 |
| complement factor B isoform 1 specific | Cfb | NP_032224.2 | 0.61 | 3.95E-08 | Not Significant | |
| cathepsin B | Ctsb | NP_031824.1 | 0.49 | 8.27E-09 | Tumors up 1.3x | 9.84E-03 |
| hemopexin | Hpx | NP_059067.2 | 1.50 | 4.34E-65 | Tumors down 5.4x | 1.23E-03 |
| rho GDP-dissociation inhibitor 1 | Arhgdia | NP_598557.3 | −1.39 | 2.54E-03 | Tumors up 1.4x | 2.24E-03 |
| afamin | Afm | NP_660128.2 | −0.31 | 0.020 | Tumors down 4x | 6.79E-03 |
| PREDICTED: aphrodisin | Obp1b, EG628991 | XP_898907.3 | −1.74 | 7.10E-19 | Not detected | |
| alpha-1B-glycoprotein | A1bg | NP_001074536.1 | −2.92 | 2.74E-09 | Not Significant | |
| apolipoprotein C-I | Apoc1 | NP_031495.1 | −0.81 | 4.20E-04 | Not Significant | |
| collagen alpha-1(I) chain | Col1a1 | NP_031768.2 | −0.98 | 1.30E-06 | Tumors up 5x | 4.08E-04 |
| platelet glycoprotein Ib alpha chain | Gp1ba | NP_034456.2 | −0.74 | 9.27E-03 | Not Significant | |
| pyruvate kinase isozymes M1/M2 | Pkm2 | NP_035229.2 | −1.10 | 2.27E-04 | Not Significant |
Value represents the the area ratio at 52 or 66 days with the lowest U-value. To see all area ratios and results, see the supplementary data.
Significance was defined as a p-value of less than 0.05.
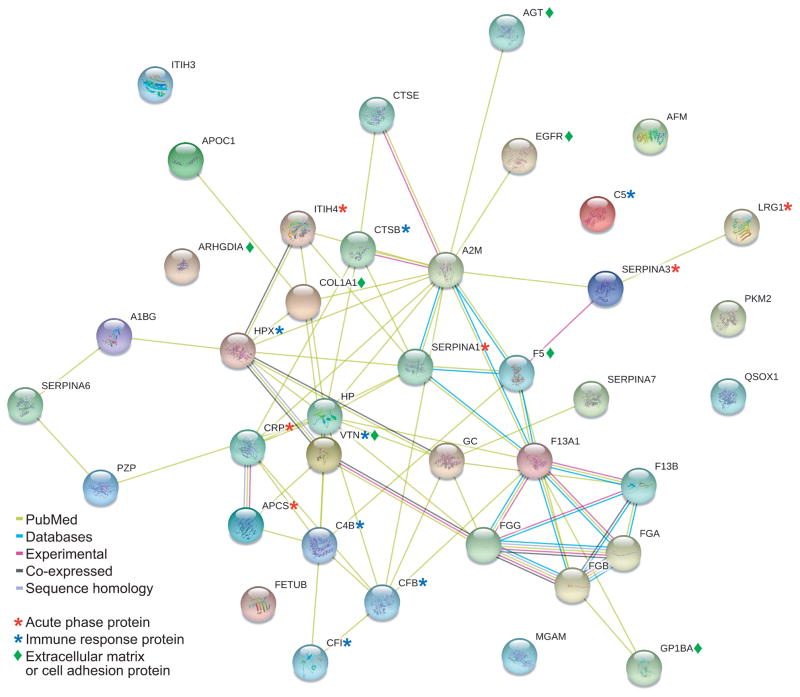
Figure 5 highlights the relationships of all of the human versions of the differentially expressed proteins in the Min mouse listed in Tables 2 and 3.32 Many differentially expressed proteins have associated connections to other proteins, and often relate to acute-phase response, innate immune response, and extracellular matrix interactions. These interactions and associated functions may play an important role in producing the Min phenotype. While multiple disease states could elicit differential protein expression in acute-phase response and innate immunity proteins, we identified other proteins that may be specific to the presence of Min intestinal tumors. For example, a protein such as maltase-glucoamylase is quite intestine-specific (see Supplementary Figure 4). While some of the proteins, such as maltase-glucoamylase, shown in Figure 5 are not connected to other proteins in this database, they may be functionally important to the intestinal neoplastic phenotype (see Discussion).
Figure 5.
String-db version 9.0 (www.string-db.org) interacting protein web for the relationships between differential expression proteins listed in Tables 2 and 3.32 This web was generated with human versions of the proteins and used a medium confidence of 0.400 or higher. Colored lines represent the type of interaction between the two proteins. Yellow-green: associated proteins are co-mentioned in PubMed abstracts, cyan: proteins are associated in curated databases, purple-pink: experimental connections, black lines: co-expressed proteins, and light blue: sequence homology. Asterisks and diamonds highlight common Gene Ontology Annotations for these proteins.
The final tier of proteins identified in this study with potential differential expression are singly identified proteins or proteins with no confirmatory reciprocal metabolic labeling data (Table 4, Supplementary Table 3). These low abundance proteins could be legitimate biomarkers given that proteins involved in cancer may not be present in high abundance, and measurements of their relative expression levels are hindered by the 1010 dynamic range of blood protein concentrations. Singly identified proteins in this category were considered only if they were upregulated two-fold or more (log2 ratio was 1.0 or higher), the peptide was unique, and both the raw MS1 and MS/MS spectra were manually validated to ensure that the computed area ratio was not an artifact. For all remaining quantitative results from the in anima study, see supplementary table 4.
TABLE 4.
Proteins differentially expressed in Min compared to wildtype with no reciprocal metabolic label information or single hits
| Protein name | Gene symbol | NCBI number | Log2 Area ratio Min/WTa | U-valuea | mRNA expression Min mouse (Tumor/Normal Epithelium)b | Microarray P-valueb |
|---|---|---|---|---|---|---|
| glycosylation-dependent cell adhesion molecule 1 | Glycam1 | NP_032160.1 | 1.08 | 2.98E-06 | Not Significant | |
| heparin cofactor 2 | Serpind1 | NP_032249.3 | 0.84 | 1.13E-03 | Tumors up 21.5x | 3.86E-06 |
| lipopolysaccharide-binding protein | Lbp | NP_032515.2 | 0.60 | 7.62E-03 | Tumors up 6.4x | 1.18E-05 |
| alpha-1-acid glycoprotein 1 | Orm1 | NP_032794.1 | 0.78 | 2.74E-05 | Not Significant | |
| peptidase inhibitor 16 | Pi16 | NP_076223.3 | 0.48 | 3.62E-04 | Not Significant | |
| delta-aminolevulinic acid dehydratase | Alad | NP_032551.3 | 1.97 | 0.048 | Tumors up 1.5x | 2.25E-04 |
| fibrinogen-like protein 1 | Fgl1 | NP_663569.2 | 1.54 | N/A | Not Significant | |
| sialic acid binding Ig-like lectin G | Siglecg | NP_766488.2 | 1.48 | N/A | Tumors down 11x | 1.25E-05 |
| Hyaluronan and proteoglycan link protein 1 | Hapln1 | NP_038528.3 | 1.54 | N/A | Tumors down 3.6x | 1.55E-03 |
| ADP/ATP translocase 2 | Slc25a5 | NP_031477.1 | 1.10 | N/A | Tumors down 2.3x | 2.21E-06 |
| Contactin-4 | Cntn4 | NP_001103219.1 | 2.69 | N/A | Not Significant | |
| Syntaxin-1A | Stx1a | NP_058081.2 | 1.32 | N/A | Tumors up 19.8x | 4.36E-06 |
| Cadherin-2 | Cdh2 | NP_031690.3 | 1.17 | N/A | Not Significant | |
| Complement C1r subcomponent-like protein | C1rl | NP_851989.3 | 1.62 | N/A | Tumors down 1.6x | 1.46E-03 |
| Fatty acid-binding protein, heart | Fabp3 | NP_034304.1 | 1.18 | N/A | Not Significant | |
| Apelin | Apln | NP_038940.1 | −1.84 | N/A | Tumors up 55.5x | 7.40E-06 |
| Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 12 protein homolog | Als2cr12 | NP_780579.1 | 1.85 | N/A | Tumors down 14.6x | 2.87E-07 |
| Gamma enolase | Eno2 | NP_038537.1 | −1.18 | N/A | Not Significant | |
| Ubiquinone biosynthesis protein COQ9, mitochondrial | Coq9 | NP_080728.1 | −1.20 | N/A | Tumors down 2.4x | 1.32E-06 |
| Dipeptidyl peptidase 4 isoform 1 and 2 | Dpp4 | NP_034204.1 | −0.68 | N/A | Not Significant | |
| Pancreatic Lipase-related protein 1 | Pnliprp1 | NP_061362.1 | −1.93 | N/A | Tumors up 85x | 1.15E-08 |
| Hypothetical protein LOC72056 | 1810055G 02Rik |
NP_082353.1 | −5.44 | N/A | Tumors up 1.5x | 1.07E-02 |
| Chymotrypsin-like elastase family member 2A | Cela2a | NP_031945.1 | −1.14 | N/A | Not Significant | |
| Isocitrate dehydrogenase [NADP], mitochondrial | Idh2 | NP_766599.2 | −1.09 | N/A | Tumors up 2x | 3.15E-04 |
| Family with sequence similarity 148, member B | C2cd4b | NP_001074783.1 | −1.24 | N/A | Not Significant | |
| Coiled-coil domain-containing protein 39 | Ccdc39 | NP_080498.1 | −1.18 | N/A | Not Significant | |
| Thrombospondin type-1 | Thbs1 | NP_035710 | −1.32 | N/A | Tumors up 6.4x | 2.64E-04 |
| Tectonic 3 | Tctn3 | NP_080536.2 | −1.85 | N/A | Tumors up 3.7x | 4.52E-04 |
| extracellular superoxide dismutase [Cu-Zn] | Sod3 | NP_035565.1 | −0.66 | 3.21E-06 | Not Significant | |
| thrombospondin-4 | Thbs4 | NP_035712.1 | 0.57 | 1.45E-03 | Tumors down 19.7x | 2.09E-04 |
| leukemia inhibitory factor receptor isoform 2 | Lifr | NP_001106857.1 | −1.35 | 4.47E-03 | Tumors up 2.7x | 1.08E-05 |
| proteasome subunit alpha type-6 | Psma6 | NP_036098.1 | −0.78 | N/A | Tumors up 1.4x | 2.99E-05 |
| lymphocyte antigen 86 | Ly86 | NP_034875.1 | −1.66 | N/A | Not Significant | |
| receptor-type tyrosine-protein phosphatase mu | Ptprm | NP_033010.2 | −1.81 | N/A | Not Significant | |
| phosphoglycerate kinase 1 | Pgk1 | NP_032854.2 | −0.68 | N/A | Not Significant | |
| phospholipase A-2-activating protein | Plaa | NP_766283.2 | −1.25 | N/A | Tumors up 1.8x | 2.46E-03 |
Value represents the the area ratio at 52 or 66 days with the lowest U-value. To see all area ratios and results, see the supplementary data. Single hit proteins have no p-value and are indicated with “N/A”.
Significance was defined as a p-value of less than 0.05.
Selected Reaction Monitoring Screen
Nine peptides derived from nine different proteins were selected for a secondary targeted SRM-MS screen. The peptides selected were chosen based on biological importance of the protein, quality of peptide in the 14N/15N discovery results, and rankings of the peptides from SRMAtlas and the mouse plasma build of PeptideAtlas.33, 34 Peptides were BLAST searched against the mouse Ref_Seq protein database to ensure uniqueness to the protein of interest. Additional considerations were made to include some proteins whose mRNA data matched the directional expression of the 14N/15N metabolic labeling study. In this way, a cross-study analysis of some proteins/gene transcripts could be done (Table 5).
Table 5.
Comparison of protein and gene expression across the three studies
| Category | Protein/gene name | 14N/15N results | mRNAa | SRM Screen |
|---|---|---|---|---|
| Same expression change in all three screens | Inter-alpha-trypsin inhibitor heavy chain H3 | + | + | + |
| coagulation factor V | + | + | + | |
| Maltase-glucoamylase | − | − | − | |
|
| ||||
| Same expression change in two out of three screens | collagen alpha-1(I) chain | − | + | − |
| Leucine-rich alpha-2-glycorprotein | + | + | ||
| Fetuin B | + | + | ||
| Cathepsin B | + | + | ||
| Haptoglobin | + | + | ||
| Complement factor I | + | + | NS | |
| serine protease inhibitor A3N | + | + | ||
| Coagulation factor XIII B | − | − | ||
| afamin | − | − | ||
|
| ||||
| Data for only two studies where opposite expression is shown | Epidermal growth factor receptor | − | NI | + |
| Hemopexin | + | − | NS | |
| alpha-1 antitrypsin 1–5 | + | − | ||
| Serum amyloid P-component | + | − | ||
| Angiotensinogen | + | − | ||
| rho GDP-dissociation inhibitor 1 | − | + | ||
|
| ||||
| Data was only achieved for the 14N/15N study, but the proteins may be biologically significant | Cathepsin E | + | NS | |
| Alpha-2-macroglobulin | + | NS | ||
| Vitronectin | + | NS | ||
| pyruvate kinase isozymes M1/M2 | − | NS | NQ | |
| Vitamin D-Binding protein | + | NI | NS | |
| Inter-alpha-trypsin inhibitor heavy chain 4, isoform 1 | + | NS | ||
All other mRNAs for the differentially expressed proteins discovered in the 14N/15N were either not significant (NS) or not identified (NI).
NS=Not significant
NI=Not identified
NQ= Identified but not quantifiable
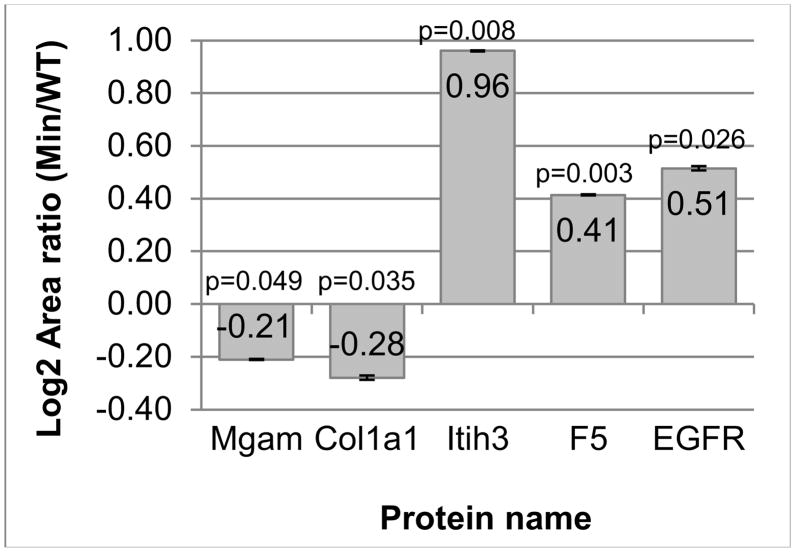
Of the 9 peptides/proteins monitored, four showed statistically significant changes in the same expression direction as the in anima metabolic labeling study (Figure 6, Supplementary Table 5). The downregulation of collagen 1 alpha-1(I) chain (COL1A1) and maltase-glucoamylase (MGAM) was confirmed. In addition, inter-alpha trypsin inhibitor, heavy chain H3 (ITIH3) and coagulation factor V (F5) were upregulated. Epidermal growth factor receptor (EGFR) was upregulated in this study, which is the opposite expression compared to the in anima metabolic labeling study. One possible explanation of this contradictory result is that since there are EGFR negative and EGFR positive colon tumors derived from the same germline genotype, the second set of mice used for this study may have shown an EGFR positive phenotype rather than an EGFR negative phenotype. Complement factor I, vitamin D binding protein, and hemopexin did not show any statistically significant changes in expression. While present, the abundance of pyruvate kinase M2 was too low for reasonable quantification.
Figure 6.
Statistically significant changes in protein expression as determined by relative quantification of SRM-MS data using stable isotope labeled peptides. Maltase-glucoamylase (MGAM), collagen 1 alpha-1(I) chain (COL1A1), inter-alpha-trypsin inhibitor heavy chain H3 (ITIH3) and coagulation factor V (F5) all matched the 14N/15N quantitative data. Epidermal growth factor receptor (EGFR) showed opposite expression compared to the 14N/15N quantitative data. The other proteins quantified either showed no statistical change or were not quantified due to too low of an abundant endogenous peptide peak. Error bars represent the average standard error of the technical replicates from all biological samples.
Quantitative microarray analysis
In addition to the quantitative proteomic analyses, a quantitative transcriptome analysis was performed comparing mRNA from tumor tissue to mRNA from normal colonic epithelium (Tables 2–5). Notably, haptoglobin, leucine-rich alpha-2 glycoprotein, and complement factor I mRNAs were upregulated quite significantly, with gene expression increased in Min tumors 63.5-fold, 110-fold, and 879-fold, respectively, over normal colonic epithelium. A concomitant increase in protein expression was also observed for these genes in the 14N/15N metabolic labeling study, although the magnitudes of increase in protein levels were not nearly as high as in the mRNA. Eight other proteins from the 14N/15N study showed trends in expression consistent with the microarray data (Table 5). Six proteins showed proteomic changes opposite to those of their mRNAs as previously reported for tumor proteins.13 Most other mRNAs showed no statistically significant change in expression in tumor tissue compared to normal colonic epithelium. As in many transcriptomic-proteomic comparisons, the directionality and level of change in mRNA expression can play a role in identifying biomarker candidates.35, 36 However, the low correlation of protein and mRNA expression in our data indicate that Min mouse mRNA expression is valuable but not sufficiently predictive for identifying candidate serum-borne protein biomarkers.
Discussion
14N/15N labeling provides beneficial internal controls
A major recurrent problem with biomarker studies is the lack of reproducibility that can arise from biological, experimental, and technical variability.37 The in anima metabolic labeling strategy employed here allows samples to be combined in equal amounts immediately upon collection. This allowed the Min and wild-type samples to be prepared and analyzed at exactly the same time at every step, thus controlling for the experimental and technical errors that can occur using many other quantitative proteomic strategies. We predicted that any change in Min protein expression compared to wild type using a 15N/14N Min-wild type labeling scheme would be reflected in the reverse metabolically labeled littermate samples. The reciprocal metabolic labeling strategy used provided a control to identify inconsistencies in protein expression due to diet-specific changes. Diet-specific changes have previously been reported using in anima metabolic labeling.38–40 In these cases, a protein preferentially incorporates one of the nitrogen isotopes regardless of which isotopes the mutant and wild type mice were fed. In the diet-specific changes observed in our study, both 14N and 15N envelopes were identified, but the protein to incorporated one of the isotopes preferentially to the other (See Supplementary Figure 3). The reciprocal metabolic labeling schema allowed us to easily differentiate between true changes and those that may be isotope related. We believe that the internal control provided by combining metabolically labeled samples at the time of collection is a benefit that controls for the few isotope-specific expression artifacts we observed.
Monitoring multiple time points revealed expression trends
The ability to collect blood at multiple time points during the life of the mouse enabled us to observe changes in the expression level of a candidate biomarker over time. Knowing whether a change trends consistently over time is an improvement over biomarker screens that make conclusions regarding biomarker efficacy based on a single time point. The proteins we observed to be the most strongly expressed in this study tended to show more consistency and statistical validity in their changes at 52 and 66 days rather than at the earliest time point of 40 days. This trend suggests that proteins changing at the later time points may be more relevant as biomarkers than those proteins that change only at the earliest time point. However, the 52 and 66 day time points are still considered early detection time points given that polyp multiplicities are low and the mice do not yet show any overt signs of illness.12, 26
Multiple quantitative screens identified proteins of biological relevance
The SRM screen and microarray data provided additional information on these putative biomarkers and allowed for quantitative analysis of the proteins/genes across multiple platforms. It should be noted that the 14N/15N, SRM, and microarray screens used C57BL/6J-ApcMin/+ x BTBR mice, but each screen used different sets of mice and the mRNA and SRM-MS screens used mice fed normal the normal breeder diet whereas the metabolically labeled set was fed exclusively the algae, Spirulina. Therefore, proteins/genes that show expression changes in the same direction across multiple types of screens provide robustness in the presence of biological variability. However, it should be cautioned that mRNA expression does not always correlate with protein expression. Below is an assessment of the biological importance derived from some of the differentially expressed proteins and gene transcripts identified in this study. A comparison of expression across all three quantitative screens is found in Table 5.
Inter-alpha-trypsin inhibitors, heavy chain H3 (ITIH3) and heavy chain 4, isoform 1
Both of these inter-alpha trypsin inhibitors were upregulated in the 14N/15N proteomics study. In addition, ITIH3 shows upregulation in the microarray and SRM screens. The upregulated inter-alpha trypsin inhibitors are involved in the covalent binding and stabilization of hyaluronic acid on the extracellular matrix.41 Hyaluronan is a large epithelial glycosaminoglycan complex known to increase in size with the growth of colonic polyps and tumors.42 ITIH3 has previously been identified as upregulated in the plasma of human gastric cancer patients and has a predicted role in the prevention of metastasis and tumor invasion activities in colon cancer.42, 43
Coagulation factor V (F5)
F5 was upregulated in all three of the quantitative studies presented in this work. Another quantitative proteomics study in the Min mouse has also shown F5 upregulation in blood.44 Coagulants such as fibrinogen (upregulated in this study), F5, and other coagulation factors have increased levels in colon cancer patients.45, 46 Specifically, F5 is most known for its association with the Factor V Leiden coagulation disease. Factor V Leiden is caused by a single nucleotide polymorphism (SNP) involving an R506Q mutation. This mutation reduces the ability of the activated protein C anticoagulant protein from binding F5. Normal interactions between activated protein C and F5 lead to the degradation of F5. However, in the absence of this interaction, F5 levels increase and cause excessive coagulation. Patients homozygous for the factor V Leiden mutation show a nearly 6-fold increased risk for colorectal cancer.46
Maltase-glucoamylase (MGAM)
All three quantitative screens showed downregulation of maltase-glucoamylase (MGAM). MGAM is an intestinal protein necessary for catalyzing the final steps in starch catabolism.47, 48 An immunohistochemical study that compared MGAM expression on human colonic epithelium to normal epithelium showed reduced expression of this enzyme.47, 48 Maltase-glucoamylase, as an intestinal protein, may provide specificity and sensitivity to blood biomarker screens for intestinal cancer. Conversely, in our previous proteomics study using colonic and intestinal tissues, MGAM was not identified or quantified13. MGAM, while quite specific to the intestine (Supplementary figure 4), is a very large protein (1827 amino acids) with a single transmembrane pass and a very small domain inside the cell.49 The brush border activity of the enzyme is localized completely external to the cell. Thus, this protein may be released into the blood. We theorize that the protein was not identified in our previous study of the tumor tissue proteome because the protein extraction could not remove the association of the protein with the plasma membrane.
Collagen-1 type 1(I) alpha 1 (COL1A1)
COL1A1 was downregulated in both the 14N/15N and SRM-MS screens while upregulated in the mRNA tissue of Min mouse colon tumors. Downregulated COL1A1 has been previously reported in cancer studies, and it is thought to play a role as part of oncogenic transformation.50 Among its many functions, COL1A1 is a positive regulator of the canonical WNT signaling pathway, the pathway that is constitutively active in early stages of colon cancer.51 COL1A1 and LRP5 expression are commonly linked in bone matrix formation and are misregulated in bone disease (Supplementary Figure 5). LRP5 is a co-receptor with the frizzled receptor in the WNT signaling pathway.
Epidermal growth factor receptor (EGFR)
EGFR was downregulated in the 14N/15N mouse study and upregulated in the SRM-MS screen. EGFR is commonly observed as upregulated in colon cancers and is the target of several colorectal cancer therapeutics.52–54 While the results of our quantitative study are seemingly contradictory, the consistency of EGFR upregulation across studies is debated.53 The frequency of upregulation ranges anywhere from 25–90%, but EGFR downregulation is also known, and EFGR-negative tumors have been reported.52–55 Because EGFR positive tumors have been implicated in poor prognosis, we believe that EGFR may be a good marker to indicate prognosis and subsequent method of treatment.56
Vitronectin
Vitronectin was upregulated in the 14N/15N study. Vitronectin has been shown to promote cell adhesion and spreading, and is indicated in tumor malignancy.57 This protein also inhibits the membrane-damaging effect of some proteins involved in the terminal cytolytic complement pathway through binding to several serpin serine protease inhibitors.58 We observed large and consistent upregulation in the Min mouse of several complement factors including complement factor B, complement C5, and complement C4-B, suggesting that vitronectin could partially mitigate the damaging effects of these upregulated proteins.
Cathepsins B and E
Among the upregulated proteins in Table 3 is cathepsin E, a gastric aspartyl protease that is found at highest levels on the mucosal producing epithelial cells of the stomach.59 It is commonly upregulated in gastric cancers. While not indicated as overexpressed in our microarray data, cathepsin E genomic transcripts are overexpressed in human serrated colonic adenomas.59 Cathepsin B protease is also showing upregulation at the mRNA level and in the 14N/15N study. Cathepsin B has been identified in elevated amounts on the surface of colon tumor cells, in plasma, and has previously been linked to negative colon cancer prognosis.44, 60 Interestingly, alpha-2-macroglobulin (Figure 4), a protease inhibitor observed to be downregulated in serum from the Min mouse, has been shown to interact with these two cathepsins (Supplementary Figure 6).61, 62 The potential negative regulation by this protease inhibitor may be related to the higher expression levels of these cathepsins in serum.
Inflammatory enzymes
Alpha-1-antitrypsin 1–5 is a protease inhibitor that helps protect tissues from the release of inflammatory enzymes and often rises in concentration during acute inflammation reactions.63 This protein has been identified at increased levels in stool samples and in serum from human colon cancer patients.63, 64 Leucine-rich alpha-2-glycoprotein (LRG1) and fetuin-B are also thought to play a role in acute phase response and inflammation.65, 66 LRG1 has shown upregulation in multiple mouse studies and has been shown to be upregulated in the plasma of human colon cancer patients.43, 44, 66, 67 Evidence of LRG1 upregulation in colon cancer across multiple studies highlights the strong relation of this protein’s expression between our study and other blood biomarker studies.
Single “hit” proteins from the 14N/15N study
While concrete conclusions cannot be drawn from the single “hit” data, there are some patterns of note in the biological functions of these proteins. Three metabolic enzymes, phosphoglycerate kinase 1, isocitrate dehydrogenase [NADP], and gamma enolase were downregulated in this dataset. It is known that pyruvate production may be reduced in colonic tumors, and that different metabolic phenotypes can be established in tumor cells.68 The tricarboxcylic acid cycle has also been shown to be downregulated in colon cancer and isocitrate dehydrogenase [NADP] is a key oxidative enzyme in this pathway.69 In addition, seven of these proteins share the Uniprot GO terms of cell adhesion as their biological process.70 This set includes upregulated hyaluronan proteoglycan link 1, a protein that helps stabilize hyaluronan proteoglycan monomers. Many of these roles map back to similar functions of differentially expressed proteins that have been identified at higher confidence levels, thus providing further confidence that these ‘single hit’ candidates may be important.
Candidate biomarkers show evidence of a whole-body response
While one intestine-specific protein was identified, organs other than the intestine secrete a significant number of the differentially expressed proteins. These proteins, often produced in the liver, do not reflect a direct secretion from the growing intestinal polyps. We confirmed this inference by comparing this study with our previous study of tumor tissue. No quantification of these presumptive liver proteins was observed among proteins found in the intestine or colon (Supplementary Table 6).
Further evidence of a bodily response distant from the tumor cells themselves can be seen when comparing microarray data of colon tumor tissue to normal epithelium. Owing to the significant number of proteins secreted from outside the small and large intestine, the proteins identified were not always expressed to any significant level in mRNAs of Min mouse tumors compared to normal epithelium (see Tables 2, 3, and 4).13 Consequently, the differential expression of tumor mRNA compared to normal colon epithelium may not fully represent the disease state. More importantly, our proteomic and transcriptomic expression data provide evidence contrary to the assertion that early cancers may not be detectable in blood due to the relatively small contribution of tumor protein secretions into blood.71 It is likely that the entire body reacts to the presence of a tumor and that proteins secreted from locations other than that of the primary lesion could significantly impact the composition of proteins in blood.
Major examples of non-tumor-specific differentially expressed proteins include the hepatically produced acute-phase and inflammatory response proteins that are upregulated in the Min mouse. Acute phase, inflammatory and immune responses have been identified as a common response to tumor presence.72, 73 While these inflammatory proteins may not be specific to intestinal cancer, we also observed secreted proteins are involved in cell adhesion, an important function related to cancer metastasis. Hyaluronan-binding proteins such as the inter-alpha-trypsin inhibitors provide vital transport of this glycosaminoglycan to growing tumors. These are just a few examples highlighted by this work that emphasize the importance of considering the systemic response to the cancer instead of focusing solely on tumor-specific proteins.
Apcmin/+ mouse biomarkers provide a framework for future biomarker studies
The in anima metabolic labeling study produced approximately 40 differentially expressed proteins in tumor-bearing Min mice compared to tumor-free wild-type mice. A subset of these proteins was quantified using SRM-MS as a secondary screen to verify their differential expression. In this case, the ApcMin/+ mouse has proven to be a useful model to explore differential protein expression as a means of intestinal cancer detection in serum. However, it does not provide tumor specificity to the colon because Min mice develop both colon and intestinal tumors. Thus the differentially expressed proteins described herein may not be specific to colon cancer or intestinal cancer. In addition, the use of Spirulina as a dietary source could have impacted the differential expression of proteins based on its differing nutrition, compared to the standard rodent food sources. The dietary difference between the in anima metabolic labeling screen and the SRM-MS screen may explain why SRM-MS did not verify differential expression of all of the 14N/15N differentially expressed proteins tested. However, some of the proteins were differentially expressed in the same direction from both proteomic studies, showing that these expression changes were robust in the presence of different diets. While we were able to show these changes in a mouse animal model, more studies would need to be done to determine whether the same changes are observed in humans. The differentially expressed proteins identified in this study are the first steps toward future biomarker screens in more specific animal models and in humans as a means for detecting intestinal cancers.
Conclusion
This study highlights the use of two quantitative proteomics techniques to identify differentially expressed proteins at early stages of intestinal neoplasia in the ApcMin/+ mouse with a comparison to transcriptome data. Among the differentially expressed proteins are those that are involved in highly relevant biological processes such as digestion, extracellular matrix remodeling, innate immunity, and acute phase response. Specifically, MGAM and COL1A1 were downregulated while ITIH3 and F5 were upregulated in both quantitative proteomic methods. Together, with the Min mouse model and these quantitative strategies, we have produced a set of candidate blood biomarkers for downstream validation of human intestinal cancer.
Supplementary Material
Acknowledgments
We thank Dr. Alexandra Shedlovsky for her valuable assistance with the experimental set-up. Linda Clipson assisted in data management and provided valuable input in the writing of this manuscript. We appreciate the mass spectrometry guidance provided by Dr. Gregory A. Barrett-Wilt and Grzegorz Sabat at the UW-Madison Biotechnology Center Mass Spectrometry Facility. Dr. Melissa Boersma and Nina Porcaro synthesized synthetic peptides at the UW-Madison Biotechnology Center Peptide Synthesis core facility. We also thank Dr. Christina Kendziorski and Jeremy D. Volkening for assistance with statistical analysis of the data. We acknowledge grant support from the following: the National Cancer Institute (R01 CA063677; WFD PI), the Institutional Clinical and Translational Research (ICTR) Grant Program to the SMPH (Marc Dressner, PI), National Institutes of Health (5 T32 GM08349 for MMI and ELH), Advanced Opportunity Fellowship through SciMed Graduate Research Scholars at University of Wisconsin-Madison (MMI), NIH Predoctoral Training Program in Genetics 5 T32 GM07133 (JKP), a Mordridge Predoctoral Fellowship (AAI), and the National Institute of Environmental Health Sciences Pre-Doctoral Training Grant T32ES007015-33 (AAI). We dedicate this manuscript to the memory of Professor Hans J. Kende.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Subramanian S, Bobashev G, Morris RJ. Modeling the cost-effectiveness of colorectal cancer screening: policy guidance based on patient preferences and compliance. Cancer Epidemiol Biomarkers Prev. 2009;18(7):1971–8. doi: 10.1158/1055-9965.EPI-09-0083. [DOI] [PubMed] [Google Scholar]
- 4.Smith RA, Cokkinides V, Brawley OW. Cancer screening in the United States, 2012: A review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J Clin. doi: 10.3322/caac.20143. [DOI] [PubMed] [Google Scholar]
- 5.Levin B, Lieberman DA, McFarland B, Andrews KS, Brooks D, Bond J, Dash C, Giardiello FM, Glick S, Johnson D, Johnson CD, Levin TR, Pickhardt PJ, Rex DK, Smith RA, Thorson A, Winawer SJ. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology. 2008;134(5):1570–95. doi: 10.1053/j.gastro.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Rex DK, Johnson DA, Anderson JC, Schoenfeld PS, Burke CA, Inadomi JM. American College of Gastroenterology guidelines for colorectal cancer screening 2009 [corrected] Am J Gastroenterol. 2009;104(3):739–50. doi: 10.1038/ajg.2009.104. [DOI] [PubMed] [Google Scholar]
- 7.Booth RA. Minimally invasive biomarkers for detection and staging of colorectal cancer. Cancer Lett. 2007;249(1):87–96. doi: 10.1016/j.canlet.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 8.Kim HJ, Yu MH, Kim H, Byun J, Lee C. Noninvasive molecular biomarkers for the detection of colorectal cancer. BMB Rep. 2008;41(10):685–92. doi: 10.5483/bmbrep.2008.41.10.685. [DOI] [PubMed] [Google Scholar]
- 9.Schneikert J, Behrens J. The canonical Wnt signalling pathway and its APC partner in colon cancer development. Gut. 2007;56(3):417–25. doi: 10.1136/gut.2006.093310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goss KH, Groden J. Biology of the adenomatous polyposis coli tumor suppressor. J Clin Oncol. 2000;18(9):1967–79. doi: 10.1200/JCO.2000.18.9.1967. [DOI] [PubMed] [Google Scholar]
- 11.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361(25):2449–60. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247(4940):322–4. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 13.Huttlin EL, Chen X, Barrett-Wilt GA, Hegeman AD, Halberg RB, Harms AC, Newton MA, Dove WF, Sussman MR. Discovery and validation of colonic tumor-associated proteins via metabolic labeling and stable isotopic dilution. Proc Natl Acad Sci U S A. 2009;106(40):17235–40. doi: 10.1073/pnas.0909282106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu CC, MacCoss MJ, Howell KE, Matthews DE, Yates JR. 3rd, Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal Chem. 2004;76(17):4951–9. doi: 10.1021/ac049208j. [DOI] [PubMed] [Google Scholar]
- 15.Surinova S, Schiess R, Huttenhain R, Cerciello F, Wollscheid B, Aebersold R. On the development of plasma protein biomarkers. J Proteome Res. 2011;10(1):5–16. doi: 10.1021/pr1008515. [DOI] [PubMed] [Google Scholar]
- 16.Sabido E, Selevsek N, Aebersold R. Mass spectrometry-based proteomics for systems biology. Curr Opin Biotechnol. 2012;23(4):591–7. doi: 10.1016/j.copbio.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Veenstra TD, Conrads TP, Hood BL, Avellino AM, Ellenbogen RG, Morrison RS. Biomarkers: mining the biofluid proteome. Mol Cell Proteomics. 2005;4(4):409–18. doi: 10.1074/mcp.M500006-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Levin Y, Jaros JA, Schwarz E, Bahn S. Multidimensional protein fractionation of blood proteins coupled to data-independent nanoLC-MS/MS analysis. J Proteomics. 2010;73(3):689–95. doi: 10.1016/j.jprot.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 19.Elschenbroich S, Kislinger T. Targeted proteomics by selected reaction monitoring mass spectrometry: applications to systems biology and biomarker discovery. Mol Biosyst. 2011;7(2):292–303. doi: 10.1039/c0mb00159g. [DOI] [PubMed] [Google Scholar]
- 20.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods. 2012;9(6):555–66. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 21.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 22.Park SK, Venable JD, Xu T, Yates JR. 3rd, A quantitative analysis software tool for mass spectrometry-based proteomics. Nat Methods. 2008;5(4):319–22. doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–8. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abbatiello SE, Mani DR, Keshishian H, Carr SA. Automated detection of inaccurate and imprecise transitions in peptide quantification by multiple reaction monitoring mass spectrometry. Clin Chem. 2010;56(2):291–305. doi: 10.1373/clinchem.2009.138420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moser AR, Dove WF, Roth KA, Gordon JI. The Min (multiple intestinal neoplasia) mutation: its effect on gut epithelial cell differentiation and interaction with a modifier system. J Cell Biol. 1992;116(6):1517–26. doi: 10.1083/jcb.116.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ball D, Rose E, Alpert E. Alpha-fetoprotein levels in normal adults. Am J Med Sci. 1992;303(3):157–9. doi: 10.1097/00000441-199203000-00004. [DOI] [PubMed] [Google Scholar]
- 28.Delmotte N, Lasaosa M, Tholey A, Heinzle E, van Dorsselaer A, Huber CG. Repeatability of peptide identifications in shotgun proteome analysis employing off-line two-dimensional chromatographic separations and ion-trap MS. J Sep Sci. 2009;32(8):1156–64. doi: 10.1002/jssc.200800615. [DOI] [PubMed] [Google Scholar]
- 29.Tabb DL, Vega-Montoto L, Rudnick PA, Variyath AM, Ham AJ, Bunk DM, Kilpatrick LE, Billheimer DD, Blackman RK, Cardasis HL, Carr SA, Clauser KR, Jaffe JD, Kowalski KA, Neubert TA, Regnier FE, Schilling B, Tegeler TJ, Wang M, Wang P, Whiteaker JR, Zimmerman LJ, Fisher SJ, Gibson BW, Kinsinger CR, Mesri M, Rodriguez H, Stein SE, Tempst P, Paulovich AG, Liebler DC, Spiegelman C. Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J Proteome Res. 2010;9(2):761–76. doi: 10.1021/pr9006365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabb DL, Fernando CG, Chambers MC. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J Proteome Res. 2007;6(2):654–61. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim S, Gupta N, Pevzner PA. Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J Proteome Res. 2008;7(8):3354–63. doi: 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database issue):D412–6. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desiere F, Deutsch EW, Nesvizhskii AI, Mallick P, King NL, Eng JK, Aderem A, Boyle R, Brunner E, Donohoe S, Fausto N, Hafen E, Hood L, Katze MG, Kennedy KA, Kregenow F, Lee H, Lin B, Martin D, Ranish JA, Rawlings DJ, Samelson LE, Shiio Y, Watts JD, Wollscheid B, Wright ME, Yan W, Yang L, Yi EC, Zhang H, Aebersold R. Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biol. 2005;6(1):R9. doi: 10.1186/gb-2004-6-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Picotti P, Rinner O, Stallmach R, Dautel F, Farrah T, Domon B, Wenschuh H, Aebersold R. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat Methods. 2010;7(1):43–6. doi: 10.1038/nmeth.1408. [DOI] [PubMed] [Google Scholar]
- 35.Gedeon T, Bokes P. Delayed Protein Synthesis Reduces the Correlation between mRNA and Protein Fluctuations. Biophys J. 2012;103(3):377–85. doi: 10.1016/j.bpj.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583(24):3966–73. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 37.Sawyers CL. The cancer biomarker problem. Nature. 2008;452(7187):548–52. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 38.Frank E, Kessler MS, Filiou MD, Zhang Y, Maccarrone G, Reckow S, Bunck M, Heumann H, Turck CW, Landgraf R, Hambsch B. Stable isotope metabolic labeling with a novel N-enriched bacteria diet for improved proteomic analyses of mouse models for psychopathologies. PLoS One. 2009;4(11):e7821. doi: 10.1371/journal.pone.0007821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Filiou MD, Webhofer C, Gormanns P, Zhang Y, Reckow S, Bisle B, Teplytska L, Frank E, Kessler MS, Maccarrone G, Landgraf R, Turck CW. The (15) N isotope effect as a means for correlating phenotypic alterations and affected pathways in a trait anxiety mouse model. Proteomics. 2012;12(15–16):2421–7. doi: 10.1002/pmic.201100673. [DOI] [PubMed] [Google Scholar]
- 40.Filiou MD, Varadarajulu J, Teplytska L, Reckow S, Maccarrone G, Turck CW. The (15) N isotope effect in Escherichia coli: A neutron can make the difference. Proteomics. 2012;12(21):3121–8. doi: 10.1002/pmic.201200209. [DOI] [PubMed] [Google Scholar]
- 41.Chen L, Mao SJ, McLean LR, Powers RW, Larsen WJ. Proteins of the inter-alpha-trypsin inhibitor family stabilize the cumulus extracellular matrix through their direct binding with hyaluronic acid. J Biol Chem. 1994;269(45):28282–7. [PubMed] [Google Scholar]
- 42.Misra S, Hascall VC, Berger FG, Markwald RR, Ghatak S. Hyaluronan, CD44, and cyclooxygenase-2 in colon cancer. Connect Tissue Res. 2008;49(3):219–24. doi: 10.1080/03008200802143356. [DOI] [PubMed] [Google Scholar]
- 43.Chong PK, Lee H, Zhou J, Liu SC, Loh MC, Wang TT, Chan SP, Smoot DT, Ashktorab H, So JB, Lim KH, Yeoh KG, Lim YP. ITIH3 is a potential biomarker for early detection of gastric cancer. J Proteome Res. 2010;9(7):3671–9. doi: 10.1021/pr100192h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hung KE, Faca V, Song K, Sarracino DA, Richard LG, Krastins B, Forrester S, Porter A, Kunin A, Mahmood U, Haab BB, Hanash SM, Kucherlapati R. Comprehensive proteome analysis of an Apc mouse model uncovers proteins associated with intestinal tumorigenesis. Cancer Prev Res (Phila) 2009;2(3):224–33. doi: 10.1158/1940-6207.CAPR-08-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paspatis GA, Sfyridaki A, Papanikolaou N, Triantafyllou K, Livadiotaki A, Kapsoritakis A, Lydataki N. Resistance to activated protein C, factor V leiden and the prothrombin G20210A variant in patients with colorectal cancer. Pathophysiol Haemost Thromb. 2002;32(1):2–7. doi: 10.1159/000057282. [DOI] [PubMed] [Google Scholar]
- 46.Vossen CY, Hoffmeister M, Chang-Claude JC, Rosendaal FR, Brenner H. Clotting factor gene polymorphisms and colorectal cancer risk. J Clin Oncol. 2011;29(13):1722–7. doi: 10.1200/JCO.2010.31.8873. [DOI] [PubMed] [Google Scholar]
- 47.Real FX, Xu M, Vila MR, de Bolos C. Intestinal brush-border-associated enzymes: co-ordinated expression in colorectal cancer. Int J Cancer. 1992;51(2):173–81. doi: 10.1002/ijc.2910510203. [DOI] [PubMed] [Google Scholar]
- 48.Young GP, Macrae FA, Gibson PR, Alexeyeff M, Whitehead RH. Brush border hydrolases in normal and neoplastic colonic epithelium. J Gastroenterol Hepatol. 1992;7(4):347–54. doi: 10.1111/j.1440-1746.1992.tb00995.x. [DOI] [PubMed] [Google Scholar]
- 49.Sim L, Quezada-Calvillo R, Sterchi EE, Nichols BL, Rose DR. Human intestinal maltase-glucoamylase: crystal structure of the N-terminal catalytic subunit and basis of inhibition and substrate specificity. J Mol Biol. 2008;375(3):782–92. doi: 10.1016/j.jmb.2007.10.069. [DOI] [PubMed] [Google Scholar]
- 50.Sengupta P, Xu Y, Wang L, Widom R, Smith BD. Collagen alpha1(I) gene (COL1A1) is repressed by RFX family. J Biol Chem. 2005;280(22):21004–14. doi: 10.1074/jbc.M413191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Medici D, Nawshad A. Type I collagen promotes epithelial-mesenchymal transition through ILK-dependent activation of NF-kappaB and LEF-1. Matrix Biol. 2010;29(3):161–5. doi: 10.1016/j.matbio.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spano JP, Fagard R, Soria JC, Rixe O, Khayat D, Milano G. Epidermal growth factor receptor signaling in colorectal cancer: preclinical data and therapeutic perspectives. Ann Oncol. 2005;16(2):189–94. doi: 10.1093/annonc/mdi057. [DOI] [PubMed] [Google Scholar]
- 53.Krasinskas AM. EGFR Signaling in Colorectal Carcinoma. Patholog Res Int. 2011;2011:932932. doi: 10.4061/2011/932932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hecht JR, Mitchell E, Neubauer MA, Burris HA, 3rd, Swanson P, Lopez T, Buchanan G, Reiner M, Gansert J, Berlin J. Lack of correlation between epidermal growth factor receptor status and response to Panitumumab monotherapy in metastatic colorectal cancer. Clin Cancer Res. 2010;16(7):2205–13. doi: 10.1158/1078-0432.CCR-09-2017. [DOI] [PubMed] [Google Scholar]
- 55.Baranovskaya S, Martin Y, Alonso S, Pisarchuk KL, Falchetti M, Dai Y, Khaldoyanidi S, Krajewski S, Novikova I, Sidorenko YS, Perucho M, Malkhosyan SR. Down-regulation of epidermal growth factor receptor by selective expansion of a 5′-end regulatory dinucleotide repeat in colon cancer with microsatellite instability. Clin Cancer Res. 2009;15(14):4531–7. doi: 10.1158/1078-0432.CCR-08-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lieto E, Ferraraccio F, Orditura M, Castellano P, Mura AL, Pinto M, Zamboli A, De Vita F, Galizia G. Expression of vascular endothelial growth factor (VEGF) and epidermal growth factor receptor (EGFR) is an independent prognostic indicator of worse outcome in gastric cancer patients. Ann Surg Oncol. 2008;15(1):69–79. doi: 10.1245/s10434-007-9596-0. [DOI] [PubMed] [Google Scholar]
- 57.Felding-Habermann B, Cheresh DA. Vitronectin and its receptors. Curr Opin Cell Biol. 1993;5(5):864–8. doi: 10.1016/0955-0674(93)90036-p. [DOI] [PubMed] [Google Scholar]
- 58.Milis L, Morris CA, Sheehan MC, Charlesworth JA, Pussell BA. Vitronectin-mediated inhibition of complement: evidence for different binding sites for C5b-7 and C9. Clin Exp Immunol. 1993;92(1):114–9. doi: 10.1111/j.1365-2249.1993.tb05956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caruso M, Moore J, Goodall GJ, Thomas M, Phillis S, Tyskin A, Cheetham G, Lerda N, Takahashi H, Ruszkiewicz A. Over-expression of cathepsin E and trefoil factor 1 in sessile serrated adenomas of the colorectum identified by gene expression analysis. Virchows Arch. 2009;454(3):291–302. doi: 10.1007/s00428-009-0731-0. [DOI] [PubMed] [Google Scholar]
- 60.Cavallo-Medved D, Dosescu J, Linebaugh BE, Sameni M, Rudy D, Sloane BF. Mutant K-ras regulates cathepsin B localization on the surface of human colorectal carcinoma cells. Neoplasia. 2003;5(6):507–19. doi: 10.1016/s1476-5586(03)80035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shibata M, Sakai H, Sakai E, Okamoto K, Nishishita K, Yasuda Y, Kato Y, Yamamoto K. Disruption of structural and functional integrity of alpha 2-macroglobulin by cathepsin E. Eur J Biochem. 2003;270(6):1189–98. doi: 10.1046/j.1432-1033.2003.03479.x. [DOI] [PubMed] [Google Scholar]
- 62.Mason RW. Interaction of lysosomal cysteine proteinases with alpha 2-macroglobulin: conclusive evidence for the endopeptidase activities of cathepsins B and H. Arch Biochem Biophys. 1989;273(2):367–74. doi: 10.1016/0003-9861(89)90495-5. [DOI] [PubMed] [Google Scholar]
- 63.Foell D, Wittkowski H, Roth J. Monitoring disease activity by stool analyses: from occult blood to molecular markers of intestinal inflammation and damage. Gut. 2009;58(6):859–68. doi: 10.1136/gut.2008.170019. [DOI] [PubMed] [Google Scholar]
- 64.Ward DG, Suggett N, Cheng Y, Wei W, Johnson H, Billingham LJ, Ismail T, Wakelam MJ, Johnson PJ, Martin A. Identification of serum biomarkers for colon cancer by proteomic analysis. Br J Cancer. 2006;94(12):1898–905. doi: 10.1038/sj.bjc.6603188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsu SJ, Nagase H, Balmain A. Identification of Fetuin-B as a member of a cystatin-like gene family on mouse chromosome 16 with tumor suppressor activity. Genome. 2004;47(5):931–46. doi: 10.1139/g04-043. [DOI] [PubMed] [Google Scholar]
- 66.Shirai R, Hirano F, Ohkura N, Ikeda K, Inoue S. Up-regulation of the expression of leucine-rich alpha(2)-glycoprotein in hepatocytes by the mediators of acute-phase response. Biochem Biophys Res Commun. 2009;382(4):776–9. doi: 10.1016/j.bbrc.2009.03.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ladd JJ, Busald T, Johnson MM, Zhang Q, Pitteri SJ, Wang H, Brenner DE, Lampe PD, Kucherlapati R, Feng Z, Prentice RL, Hanash SM. Increased plasma levels of the APC-interacting protein MAPRE1, LRG1, and IGFBP2 preceding a diagnosis of colorectal cancer in women. Cancer Prev Res (Phila) 2012;5(4):655–64. doi: 10.1158/1940-6207.CAPR-11-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thangaraju M, Carswell KN, Prasad PD, Ganapathy V. Colon cancer cells maintain low levels of pyruvate to avoid cell death caused by inhibition of HDAC1/HDAC3. Biochem J. 2009;417(1):379–89. doi: 10.1042/BJ20081132. [DOI] [PubMed] [Google Scholar]
- 69.Bi X, Lin Q, Foo TW, Joshi S, You T, Shen HM, Ong CN, Cheah PY, Eu KW, Hew CL. Proteomic analysis of colorectal cancer reveals alterations in metabolic pathways: mechanism of tumorigenesis. Mol Cell Proteomics. 2006;5(6):1119–30. doi: 10.1074/mcp.M500432-MCP200. [DOI] [PubMed] [Google Scholar]
- 70.Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40(Database issue):D71–5. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hori SS, Gambhir SS. Mathematical model identifies blood biomarker-based early cancer detection strategies and limitations. Sci Transl Med. 2011;3(109):109ra116. doi: 10.1126/scitranslmed.3003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454(7203):436–44. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 73.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.