Figure 3.
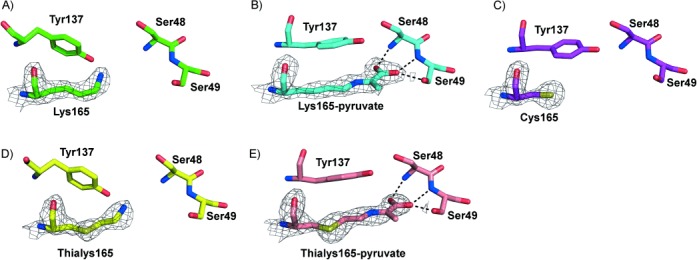
X-ray crystal structures of active-site residues of wild-type and chemically modified S. aureus NAL. A) Wild-type apoenzyme (4AHP). B) The Schiff base of Lys165 with pyruvate (4AH7). The pyruvate carboxylate group interacts with the backbone amides of Ser48 and Ser49 and the side-chain hydroxyl group of Ser49, identical to the interactions with Ser47 and Thr48 previously seen in the pyruvate complex of the E. coli enzyme.[29] C) The structure of the active site of the K165C enzyme (4AHQ). D) The active site of the γ-thialysine-165 enzyme (4AHO). E) The structure of the Schiff base formed between pyruvate and γ-thialysine at position 165 in the chemically modified enzyme (4AMA). The carboxylate makes the same interactions as for the wild-type S. aureus enzyme. In all cases the structure of subunit A of the tetramer is illustrated and electron density around residue 165 is mapped at the 1.4 σ level. In each case the atoms are coloured by atom type with the carbon atoms coloured green in the wild-type structure, cyan in the wild-type pyruvate complex, purple in the K165C structure, yellow in the thialysine-containing NAL and salmon in the thialysine-NAL–pyruvate complex.
