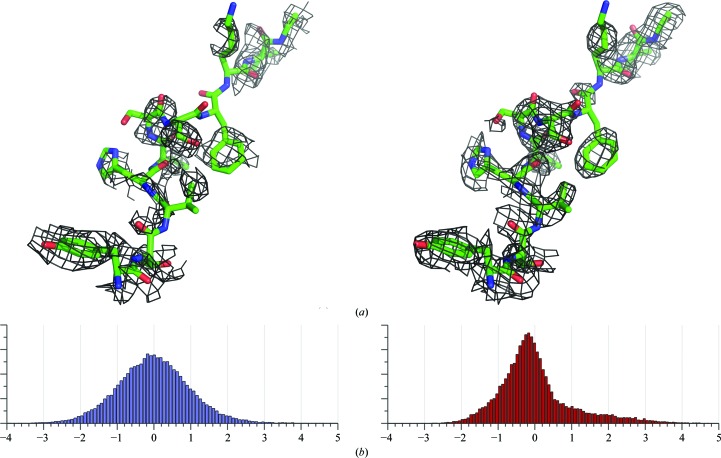
Figure 1.
Two key ideas exploited in the implementation of the method. (a) A comparison of two density-modified maps generated from SAD data for the gene V protein: the first map was derived from a reflection set with the original centroid phases (ϕB), while the second map was derived by assigning correct phases (from the PDB model) to the 100 strongest reflections from the same data set. The map correlation of the second map was significantly improved from 0.45 to 0.66. (b) A comparison of two electron-density histograms: the histogram on the left was generated from the electron-density map calculated using the centroid phases (ϕB) resulting in a small value of map skewness (skew = 0.22; see equation 1), while the histogram on the right was generated from the map calculated using the correct phases (ϕC), resulting in a large value of map skewness (skew = 1.11).