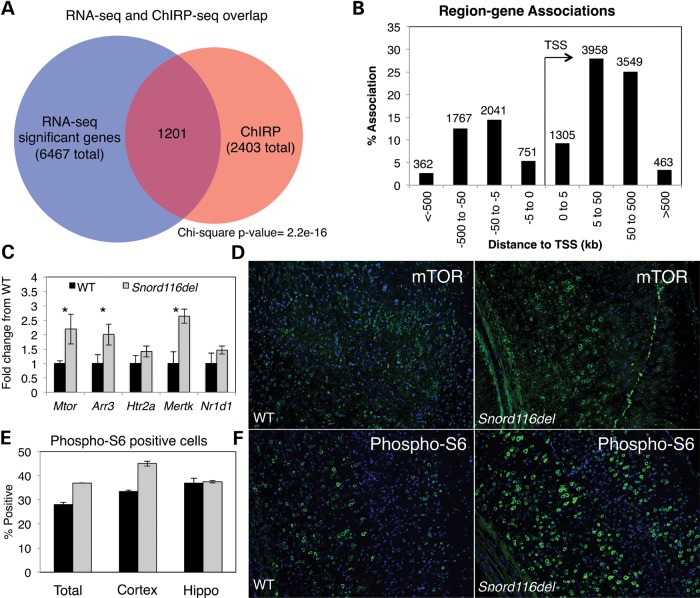
Figure 4.
Combined analysis of 116HG ChIRP-seq with RNA-seq of WT versus Snord116del cortex reveals up-regulation of 116HG bound genes including Mtor. (A) Genes associated with 116HG-specific ChIRP peaks overlapped significantly with genes showing transcript level changes by RNA-seq. (B) 116HG binding sites show clustering 5–500 kb up or downstream from transcription start sites. (C) qRT-PCR confirms significant up-regulation of 3/5 genes identified by RNA-seq: Mtor (P = 0.042), Arr3 (P = 0.049), Mertk (P = 0.007) in a larger set of WT (n = 6) and Snord116del (n = 12) mouse cortex. (D) Immunofluorescence staining of WT and Snord116del prefrontal cortex for mTOR showing increased expression in Snord116del. (E) Laser scanning cytometry quantitation of phospho-S6 staining of WT (black, n = 3) compared with Snord116del (grey, n = 3) for total brain slice, cortex or hippocampus. (F) Immunofluorescence staining of WT and Snord116del cortex for phospho-S6.