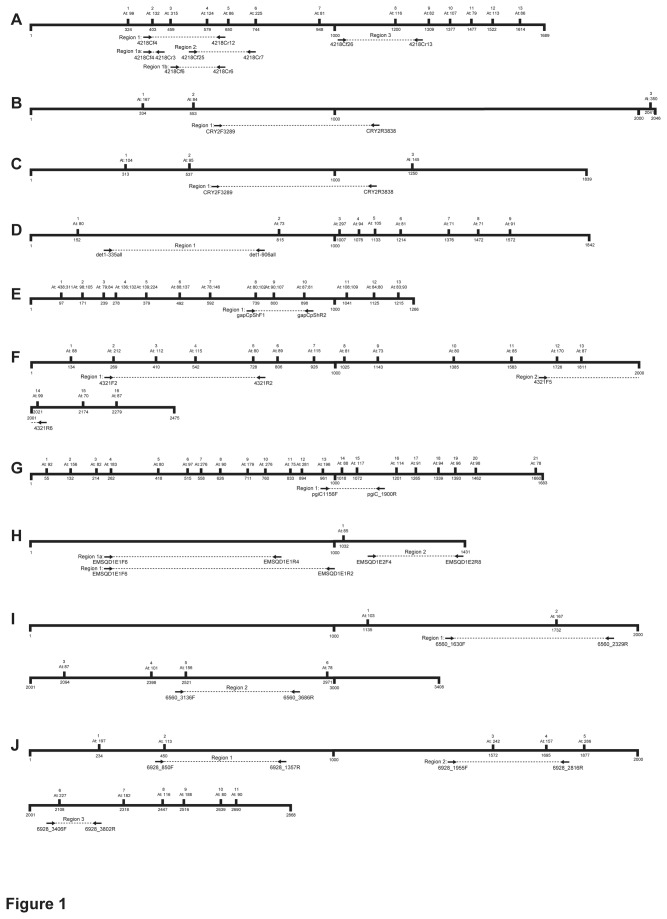
Figure 1. Schematic diagrams of the ten nuclear genes for which we developed fern-specific primers.
(A) ApPEFP_C; (B) CRY2; (C) CRY4; (D) DET1; (E) gapCpSh; (F) IBR3; (G) pgiC; (H) SQD1; (I) TPLATE; (J) transducin. Each subset of the figure represents one protein-coding locus, using the most closely related Arabidopsis thaliana homolog as the template. The coding sequence is measured (in base pairs) along the bottom of the thickened horizontal line, with each locus wrapping onto a new line every 2000 base pairs, when necessary. Intron location, number, and length (in base pairs in Arabidopsis) are given above the line. Also shown below the line are the priming locations for each of the markers we developed. For gapCpSh, intron locations are based on Arabidopsis gapCp1: the first two exons of Arabidopsis gapCp2 are each one codon shorter than in gapCp1.