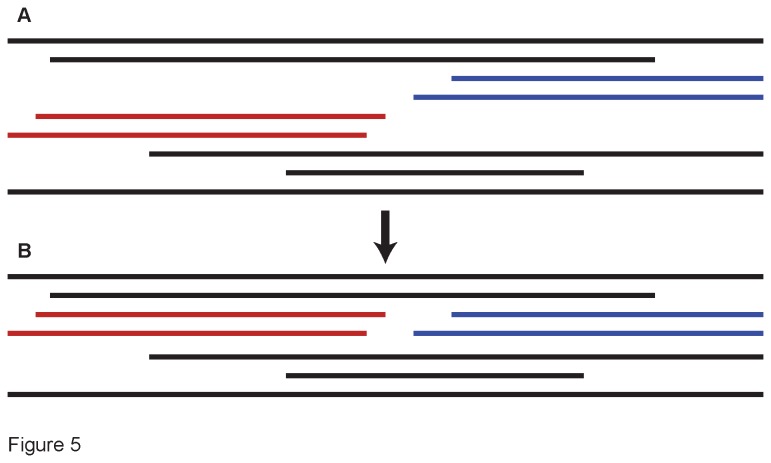
Figure 5. Example of our sequence-merging protocol.
(A) In this schematic of a transcriptome alignment, aligned sequence fragments are indicated by the horizontal bars. Included are four fragments (colored) from our focal accession, which group together in the maximum parsimony tree. However, the two fragments from the 5’ end of the protein (in red) have some base pair conflicts with each other, as do the fragments from the 3’ end (in blue). Since the two sets of fragments do not overlap, and they group in the same area of the MP tree, it is not possible to determine which 5’ fragment belongs with which 3’ one. In this case we merged the sequences arbitrarily (B). The resulting alignment retains the full nucleotide data for primer-design purposes, but the relationships at the tips of the tree may be erroneous due to the two potentially chimaeric sequences.