Figure 4.
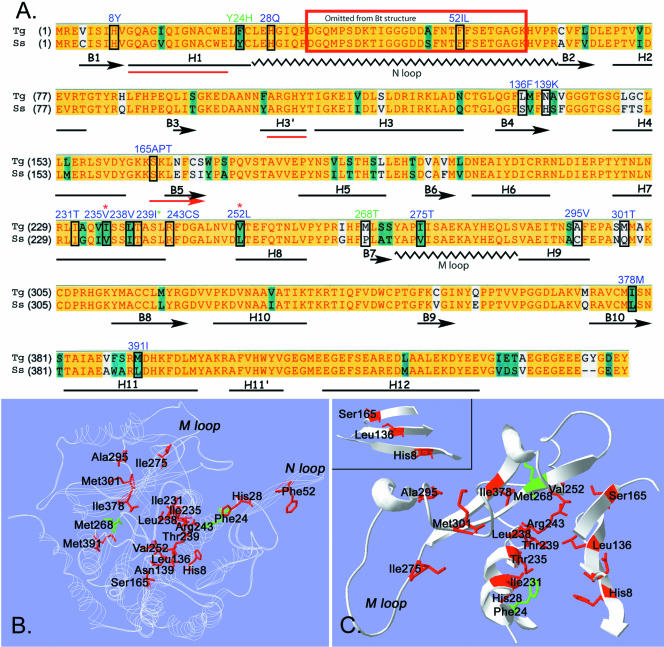
(A) Clustal alignment of oryzalin-sensitive Toxoplasma (Tg) and oryzalin-insensitive pig (S. scrofa, Ss) α-tubulins. A yellow background represents amino acid identity, teal denotes residue similarity, and white indicates nonconserved residues. The distribution of α-helices, β-strands, and coils in bovine α-tubulin (black, below Clustal sequence) were adapted from Lowe et al. (2001) who solved the structure of bovine tubulin, but mapped it onto the amino acid sequence of pig tubulin. The three red lines (H1, H3′, and B5) indicate regions where the predicted secondary structure of Toxoplasma α-tubulin deviates from the bovine secondary structure. The boxed residues are mutated in the individual oryzalin-resistant lines. The blue-labeled residues were identified in Toxoplasma oryzalin-resistant lines in this study. Three dinitroaniline resistance mutations (two labeled in green and one with a green asterisk) were identified in previous work in Chlamydomonas and Eleusine. (B and C) The amino acid sequence of Toxoplasma α-tubulin was fit to the structure of bovine α-tubulin by using Swiss-Model automated comparative protein modeling. B shows a ribbon model of Toxoplasma α-tubulin (white). The residues that are mutated to confer oryzalin resistance are colored red. The plant mutations at residues 24 and 268 are colored green. The position of the N loop containing the Phe52 mutations is not necessarily accurate because this area was disordered in structural studies and was eliminated from the final model. C shows cut-away views of some of the α-tubulin residues that are mutated to confer oryzalin resistance. Many residues are in striking proximity and show specific orientations. For example, residues Ile231, Ile235, and Thr239 all occur on the same face of α-helix 7. Leu238, Arg243, and Val252 cluster in the same general area. The inset shows a β-sheet formed by β-strands 1, 4, and 5. The mutated residues His8, Leu136, and Ser165 are in a linear pattern across the β-sheet.