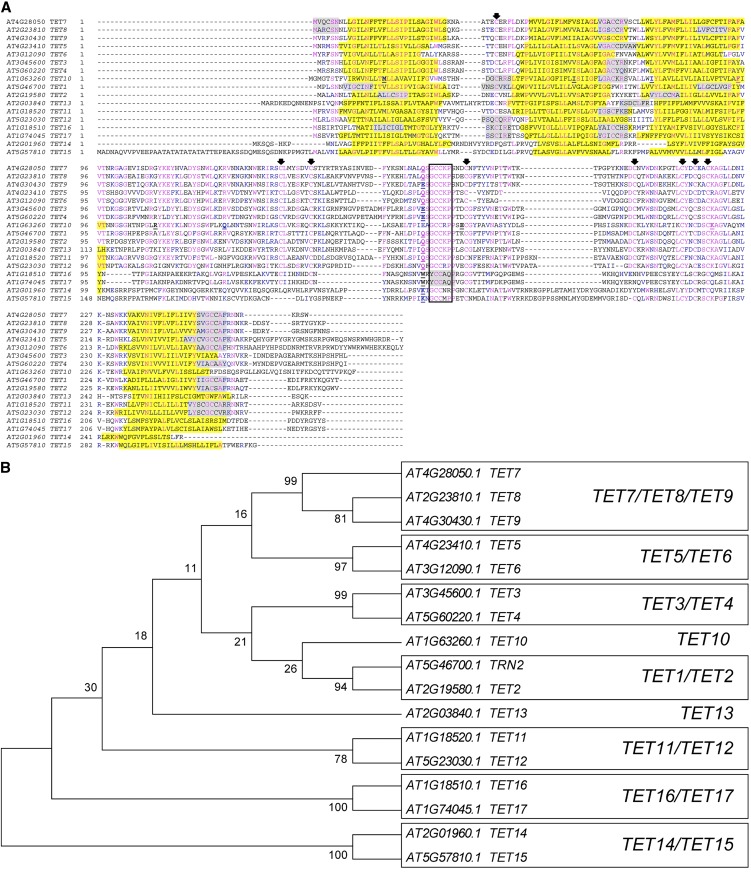
Figure 1.
Phylogenetic analysis of Arabidopsis tetraspanins. A, Amino acid sequence alignment of Arabidopsis tetraspanins. Identical residues are highlighted in blue and conserved residues in magenta. The transmembrane domains are shaded in yellow and predicted palmitoylation sites in gray. Intron positions are indicated by an underlined bold letter. Conserved Cys residues are indicated by arrows. Unfilled black box indicates the GCCK/RP motif conserved among all Viridiplantae tetraspanins. B, Unrooted neighbor-joining tree of Arabidopsis tetraspanins. The evolutionary distances were computed using the Dayhoff matrix-based method using MEGA5. The bootstrap consensus tree was inferred from 1,000 replicates. Numbers above branches indicate bootstrap percentage values for a particular branch. Boxes represent clades supported by bootstrap values above 50%. [See online article for color version of this figure.]