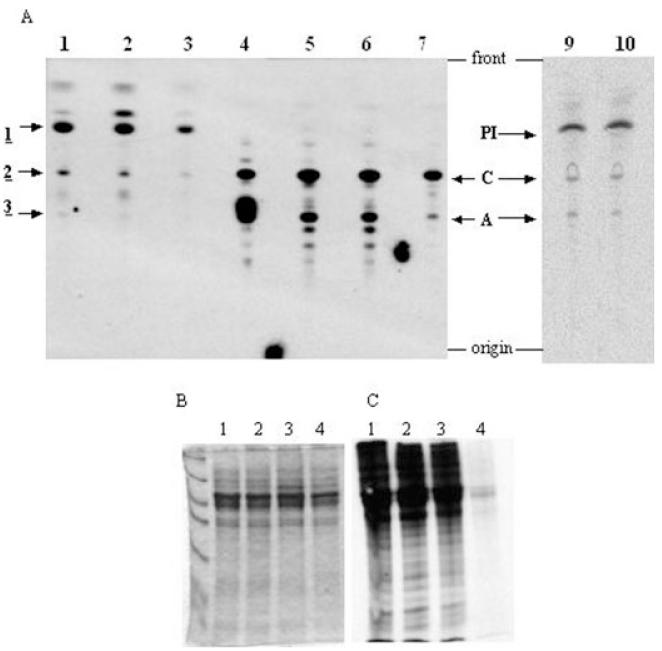
Fig. 6.
Phenotype analysis of the INO1 conditional knockout by in vivo labelling. A. Lipids from in vivo labelling with either [3H]-glucose, [3H]-mannose or [3H]-inositol were extracted, desalted and separated by HPTLC; radiolabelled lipids were detected by fluorography. Lipids from [3H]-glucose labelling are in lane 1 wild-type cells, lane 2 INO1 conditional knockout cells grown in the presence of tetracycline and lane 3 INO1 conditional knockout cells grown in the absence of tetracycline. Lipids from [3H]-mannose labelling are in lane 5 wild-type cells, lane 6 INO1 conditional knockout cells grown in the presence of tetracycline and lane 7 INO1 conditional knockout cells grown in the absence of tetracycline. Lane 4 contains glycolipids A and C standards. Lipids from [3H]-inositol labelling are in lane 9 wild-type cells and lane 10 INO1 conditional knockout cells grown in the absence of tetracycline.
B and C. Cells were labelled with 35S-methionine, and proteins were separated by SDS-PAGE and detected by either Coomassie blue staining or fluorography. Lane 1, wild-type cells; lane 2, TbINO1 conditional double knockout cells grown in the presence of tetracycline; lane 3, TbINO1 conditional double knockout cells grown in the absence of tetracycline; and lane 4 wild-type cells pre-incubated for 5 min with 60 μg ml−1 cycloheximide prior to labelling.