Abstract
Tissue-engineering therapies have shown early success in the clinic, however, the cell–biomaterial interactions that result in successful outcomes are not yet well understood and are difficult to observe. Here we describe a method for visualizing bone formation within a tissue-engineered construct in vivo, at a single-cell resolution, and in situ in three dimensions using two-photon microscopy. First, two-photon microscopy and histological perspectives were spatially linked using fluorescent reporters for cells in the skeletal lineage. In the process, the tissue microenvironment that precedes a repair-focused study was described. The distribution and organization of type I collagen in the calvarial microenvironment was also described using its second harmonic signal. Second, this platform was used to observe in vivo, for the first time, host cells, donor cells, scaffold, and new bone formation within cell-seeded constructs in a bone defect. We examined constructs during bone repair 4 and 6 weeks after implantation. New bone formed on scaffolds, primarily by donor cells. Host cells formed a new periosteal layer that covered the implant. Scaffold resorption appeared to be site specific, where areas near the top were removed and deeper areas were completely embedded in new mineral. Visualizing the in vivo progression of the cell and scaffold microenvironment will contribute to our understanding of tissue-engineered regeneration and should lead to the development of more streamlined and therapeutically powerful approaches.
Introduction
Regeneration strategies employing a scaffold material combined with cells have shown early success in the clinic,1,2 with others rapidly approaching this stage.3–5 However, the cellular mechanisms underlying regeneration are not yet well understood.6,7 Describing the progression of host–donor–biomaterial interactions behind a successful or unsuccessful therapy is still a difficult process, yet is central to the safe transfer of basic research to a clinical setting. For example, what is the fate of seeded cells? How do these cells induce or conduct repair? What are the cell and scaffold interactions governing a successful or unsuccessful therapy? To understand the impact of a given regenerative strategy on the resultant cellular interactions, it is first necessary to observe them.
In vivo microscopy has dramatically contributed to our understanding of development,8 neuroscience,9–12 immunology,13–18 and stem cell dynamics.19–21 Much of this work relies on two-photon microscopy, the theoretical foundation of which was conceived in 193122 and put into practice in 1990.23 Due to a longer excitation wavelength than conventional fluorescence microscopy, two-photon microscopy provides a deeper tissue penetration, allowing observation of cellular dynamics in situ and in vivo.24 Additionally, a longer excitation wavelength enables collagen to be visualized in vivo without the use of an additional stain due to its generation of a second harmonic signal.25–28 More recently, in vivo fluorescence imaging and two-photon microscopy have been applied to tissue-engineering applications.29,30 Concurrently, X-ray tomography31,32 and whole body fluorescence measurements33 have also provided insights into the cellular interactions governing regeneration in live animals. However, at present, whole body fluorescence imaging does not facilitate single-cell resolution and X-ray tomography does not allow integration with fluorescent reporter genes.34 Fluorescent reporter genes have allowed tracking of lineage-specific cell populations in vitro or in vivo without additional stains. Histology can provide a large field of view of reporter gene activity within a tissue of interest, however, the process compromises the viability of the tissue and the microenvironment can be physically disturbed due to sectioning. Therefore, two-photon microscopy has the distinct advantage of visualizing tissue at a single-cell resolution in vivo and in situ in three dimensions.21 By providing a description of a tissue microenvironment across multiple time points in the same animal, two-photon microscopy provides a more continuous perspective than has previously been shown using different animals at each time point.
This article describes the establishment of a two-photon microscopy platform to examine cell–scaffold-based therapies for bone regeneration in vivo. To establish a live animal imaging platform, experiments were conducted in two steps: (1) to describe the initial calvarial microenvironment before injury repair, and (2) to observe the interaction of cells, scaffold, and new bone during tissue-engineered regeneration in a calvarial defect model. To provide an unambiguous description of the spatial context in which the three-dimensional (3D) two-photon data reside, the microenvironment of calvarial bone is described starting with two-dimensional (2D) techniques of histology and stereomicroscopy, and then with 3D two-photon live animal microscopy. Each imaging technique provides a different perspective; stereomicroscopy provides a global view, histology a cross section, and two-photon a 3D volume.
We have previously employed a calvarial defect model of bone repair in combination with transgenic mice carrying reporters for cells in the skeletal lineage.35–37 To assess the cellular interactions within our cell–scaffold system, a murine critical size calvarial defect model was used because the calvarium is thin and reasonably accessible to a microscope objective. Tissue-engineered bone formation was observed in animals at 4 and 6 weeks after implantation of a scaffold and donor bone marrow stromal cells (BMSCs). Two-photon microscopy during tissue-engineered bone formation enabled visualization of host cells, donor cells, scaffold, and type I collagen at two time points, in vivo and in situ.
Materials and Methods
Live animal imaging
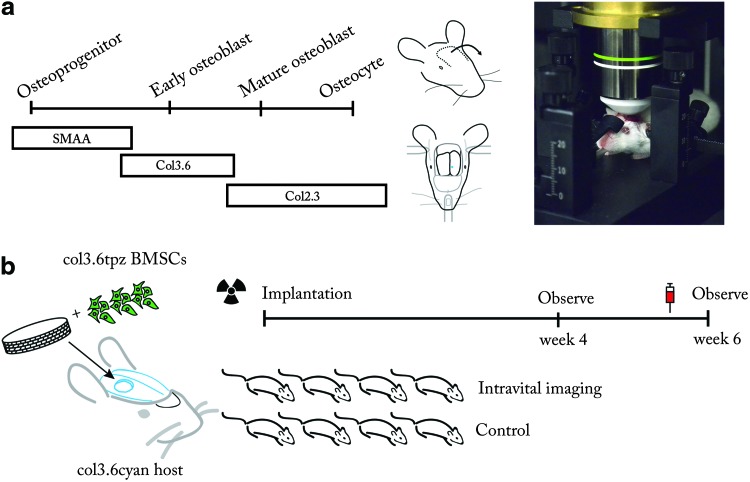
A bone mineralization label (Alizarin complexone) was injected into the tail vein of the mouse to be examined 1 day before an imaging session. On the day of imaging, the mouse was anesthetized with a ketamine (135 mg/kg)–xylazine (15 mg/kg) blend. If the vasculature was to be visualized, 100 μL of rhodamine-labeled dextrans (50 μg/μL) were injected into the tail vein. The skin above the calvarium was shaved and a u-shaped incision was made (Fig. 1a). The skin flap was sutured to an area above the nose and the animal was placed in a stereotaxic frame (Mouse and Neonatal Rat Adaptor #51625, Stoelting). The head was stabilized at three points by two blunt ear bars and a bite bar (Fig. 1a). The animal was then placed beneath a two-photon microscope (Ultima IV; Prairie Technologies). Adjusting the height of the ear bars leveled the exposed calvarium. The calvarium was imaged with a water immersion objective (XLUMPlanFL 20x/0.95W; Olympus) in sterile phosphate-buffered saline (PBS). To acquire a stack of images through the depth of the calvarium, a 459 by 459-μm area was scanned in the x-y plane, while the z depth was increased by 1-μm increments, controlled by the acquisition software (Prairie Viewer; Prairie Technologies). All fluorophores were excited at 900 nm and data were collected in four channels with the following filter limits: channel 4, 435–485 nm; channel 3, 500–550 nm; channel 2, 570–620 nm; and channel 1, 640–680 nm. When excited at 900 nm, type I collagen emits a 450 nm second harmonic; this signal was detected in channel 4. Cyan fluorescent protein-expressing osteoblasts (Col3.6cyan) were detected in channels 3 and 4. Yellow fluorescent protein-expressing osteoblasts (Col3.6topaz) were detected in channels 2 and 3. Emerald green fluorescent protein-expressing osteoprogenitors, osteoblasts, and osteocytes (SMAAemd, Col2.3emd) were detected in channel 3. The scaffold (Healos, DePuy) was autofluorescent in all channels. Rhodamine-labeled vasculature and Alizarin complexone mineral label were detected in channel 1, but used separately in the current study. After the completion of an experiment, z-stacks were reconstructed in three dimensions using the 3D viewer plugin38 for FIJI.39 This study used 3-month-old B6 (SMAAemd) and CD1 (Col3.6topaz, Col3.6cyan, Col2.3emd) mice (Fig. 1a). All procedures in this study were approved by the UConn Health Center Institutional Animal Care and Use Committee (IACUC).
FIG. 1.
Examination of skeletal cells in native and tissue-engineered bone with live animal microscopy. (a) Reporters of the skeletal lineage used in this study, surgical procedure, and animal stabilization on the microscope. Left, skeletal lineage reporters used in this study. Middle, the calvarium was exposed by incision and the skin flap was sutured down. The stereotaxic frame stabilizes the head by three-point fixation at the ear canals and front teeth. Right, animal positioned under the objective lens of the two-photon microscope. (b) Observing tissue-engineered bone regeneration. Col3.6topaz bone marrow stromal cells (BMSCs) were seeded onto a collagen-hydroxyapatite scaffold and implanted into a Col3.6cyan host. Host animals were irradiated and given a bone marrow transplant to prevent immunorejection of donor cells. At both 4 and 6 weeks later, animals in the imaging group were examined by two-photon microscopy. For comparison, the control group did not undergo imaging surgery. To visualize the mineral surface, Alizarin complexone was injected into both groups 1 day before sacrifice. Color images available online at www.liebertpub.com/tec
Calvarial defect model of tissue-engineered bone
BMSCs were isolated from the femur and tibia of CD1 wild-type and Col3.6topaz animals for bone marrow rescue following irradiation and scaffold seeding, respectively. Cells were plated in α-MEM (Gibco) containing 10% FBS and 1% penicillin–streptomycin. Following isolation, cells were allowed to attach in an incubator at 5% CO2, ambient O2, and 37°C. After 7 days of culture, Col3.6topaz cells were trypsinized and seeded onto a collagen–hydroxyapatite scaffold (Healos) at a density of 1.0×106 cells/scaffold at the time of implantation. To prevent immunorejection of the cell–scaffold implant, Col3.6cyan host animals underwent 900 rads of irradiation to ablate the bone marrow 1 day before implantation. Host animals subsequently received a bone marrow transplant of 1.5×106 cells from wild-type CD1 animals via retro-orbital sinus injection. Animals were anesthetized with a ketamine (135 mg/kg)–xylazine (15 mg/kg) blend and a 3.5-mm-diameter defect was introduced in the right parietal lobe using a Dremel MultiPro drill with a trephine bit. Extreme care was taken to prevent damage to the dura mater beneath the calvarium. After the scaffold–cell construct was placed in the defect, the incision was closed with resorbable sutures and the animals were given analgesic (bupronephrine, 0.08 mg/kg). To enable imaging at 4 and 6 weeks, animals were anesthetized, and an incision was made above the defect area. To assess the effect of imaging surgery on bone formation, two groups of animals were used. One group (n=4) underwent surgery to expose the calvarium and was subsequently imaged with in vivo two-photon microscopy at 4 and 6 weeks postimplantation. The other group (n=4), acting as a control, did not undergo surgery or imaging. Both groups were sacrificed at 6 weeks postimplantation and examined with histology and X-ray imaging.
Histology, stereomicroscopy, and X-ray imaging
Concluding a two-photon experiment, animals were sacrificed by CO2 asphyxiation. The calvaria were dissected, placed in 10% formalin at 4°C for 1 day, and then in a 30% sucrose solution overnight. Photographs and X-ray images (LX60; Faxitron) were taken. The calvaria were then placed in a sucrose solution in a culture dish beneath the stereomicroscope objective (Lumar V12; Zeiss). Images were acquired on the stereomicroscope using a digital camera (Axiocam; Zeiss) and filter set (49002, 31002 and 31043; Chroma). Images were acquired at different locations using stage automation; these resultant images were stitched together with acquisition software (AxioVision; Zeiss) to capture a large field of view. Following stereomicrosopy, each calvarium was covered with an embedding medium (Cryomatrix; Termo Shandon) and affixed to an aluminum stage for cryosectioning. Thin sections (7 μm) of tissue were transferred via tape (Cryofilm; Section-Lab) to a plastic slide. Sections were then rinsed three times in PBS, followed by distilled water, and placed on a glass slide. A 50% glycerol solution was applied to the section and a glass coverslip was placed on top. Images of finished slides were acquired with a fluorescent microscope (ImagerZ1; Zeiss) equipped with a digital camera (Axiocam; Zeiss) and filter set (49002 and 31002; Chroma).
Image analysis
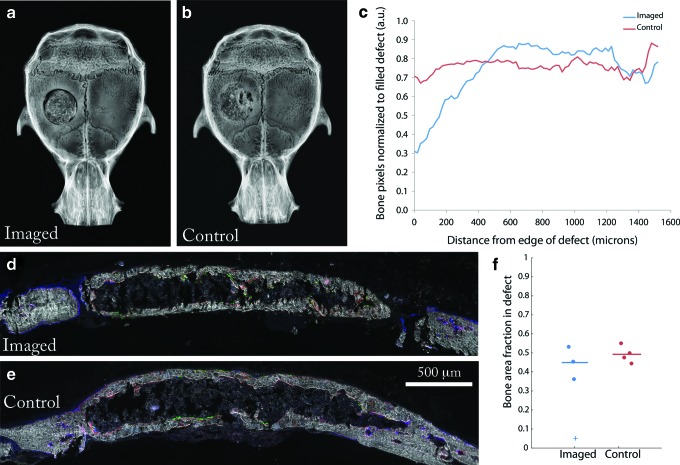
To quantify integration of host bone with the edge of the scaffold, a Euclidean distance analysis19 was performed on X-ray images of calvaria in dorsal view from both groups using FIJI.39 First, the inside edge of the host bone was selected manually with an elliptical region of interest (ROI) and saved for the following step. The entire image was then cleared to white (8-bit grayscale value of 255) and the outline of the saved ROI was drawn in black (8-bit grayscale value of 0) on the white image. A Euclidean distance map (EDM) was produced from this image, coding each white pixel a grayscale value corresponding to its distance in pixels from the black ROI representing the edge of the defect. A duplicate of the original X-ray image was then converted to a binary image (0 or 255 only) by setting pixels above a threshold value to 0 and below to 255, for bone and nonbone pixels, respectively. To combine the distance information from the EDM with the location of bone pixels from the binary image, the pixel values between the two images were compared, and the maximum value was chosen to form a composite image. Calculating a histogram of the composite image yielded the number of bone pixels as a function of pixel distance from the edge of the host bone. White background pixels (255) in the histogram were excluded from the analysis. The same procedure was repeated with the entire defect area set to 0, representing the defect as theoretically filled with bone, that is, perfect host-implant integration. To normalize these data, the histogram derived from the X-ray composite was then divided by the histogram representing perfect host-implant integration. For each group, the mean value of bone pixels was plotted as a function of distance from the edge of the defect. The pixel distance was converted to microns using the spatial calibration for the magnification used.
The area fraction of bone in the defect area was measured from histological sections using the following method. The defect was cropped with a rectangular ROI and pixels were classified as either bone or not bone using the WEKA trainable segmentation plugin40 for FIJI. The user first identified a selection of pixels belonging to each class (bone, nonbone) and the computer classified the entire image. The misclassification error was less than 0.2% for images examined by this method. The classified image was then output as a binary image. An ROI that had been manually drawn around the defect area was applied to the classified image. A histogram was calculated, resulting in the number of bone pixels and total pixels in the ROI. Dividing the number of bone pixels by the total pixels in the ROI gave the bone area fraction in the defect.
To determine the thickness of new mineral on the host bone, an ROI was manually defined around a second harmonic signal indicative of collagen within mineralized tissue (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/tec). Our rationale for classifying a second harmonic signal as collagen within the mineralized tissue is based on observing the morphology of the second harmonic signal in native bone. We also observed this morphology in areas that appeared to be new bone formation in the defect model. When the mineral label (Alizarin complexone) was administered, the label localized to the surfaces of this second harmonic signal, indicating that such a morphology was due to the collagen within the mineralized tissue. After region selection, the contents of the ROI were set to 255 and processed with the local thickness plugin41 implemented in FIJI. This plugin calculates the thickness of a 2D region using a circle filling algorithm, coding each pixel a value corresponding to the diameter of the circle it is contained by. A histogram of the image provided the mean pixel value corresponding to the mean thickness of new mineral on the host bone. New mineral on the scaffold surface was determined using the WEKA trainable segmentation plugin to classify pixels as either scaffold or new mineral. The resultant image was used to calculate the mean thickness using the local thickness plugin. The mineral apposition rate (MAR) was calculated by dividing the mean thickness of mineral on either the scaffold or host bone by the time since implantation.
Results
Surveying the calvarial microenvironment in two and three dimensions
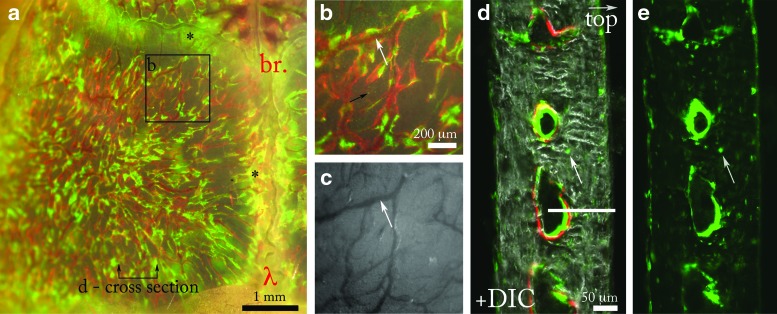
To orient 3D two-photon data within the context of the surrounding tissue and aid interpretation of two-photon data when viewed alone, a Col2.3emd42 transgenic animal was examined, with two-photon microscopy, stereomicroscopy, and histological sectioning, in this order. Here we will describe the 2D data obtained by stereomicroscopy and histology first, followed by the 3D two-photon data. The Col2.3emd reporter shows osteocytes and osteoblasts via the emerald green fluorescent protein and the mineral surface was labeled with Alizarin complexone (red). Starting with stereomicroscopy, a bird's eye view of the calvarium-contained osteoblasts distributed throughout marrow spaces in the parietal bone and suture lines (Fig. 2a). Osteoblasts colocalized with the mineralization label covering the endosteal surface (Fig. 2b). Osteocytes could be seen embedded in the cortical bone (Fig. 2b, black arrow). Blood vessels reside within the marrow spaces of the parietal bone (Figs. 2b, c, white arrows are placed at identical locations). Taken together, this data represent the repeating motif of parietal bone in the calvarium.
FIG. 2.
Osteoblasts, osteocytes, and mineralization in the mouse calvarium. (a) View of the left parietal lobe of a calvarium from a Col2.3emd mouse showing osteoblasts (green) and mineral surface labeled with Alizarin complexone (red). The green, red, and brightfield channels are overlaid to show the colocalization of the osteoblasts and new bone label. The sagittal and coronal suture lines also contain osteoblasts, indicated by the black asterisks (*). The horizontal line indicates the approximate location of the cross-sectional view shown in (d). The anatomical locations lambda and bregma are labeled λ and br., respectively. (b) Inset from (a) shows a magnified view of the marrow spaces located in the parietal bone. Blood vessels observed with brightfield optics (white arrow in c) localize to the inside of the marrow spaces (white arrow in b, same location as c). (c) Brightfield channel of (b) showing the organization of vasculature (white arrow). (d) Histological section of (a) showing Col2.3emd osteoblasts and osteocytes (white arrow) in the marrow spaces overlaid with a differential interference contrast channel showing cortical bone. Mineralization label (red) is located on the surface of endosteal marrow spaces. The white line indicates the 139-μm total depth of the two-photon z scan shown in Figure 3a. (e) Emerald signal of (d) showing osteocytes (white arrow) in the cortical phase. Color images available online at www.liebertpub.com/tec
Histology of the 200-μm-thick calvarium was used to examine the same tissue in a cross section (Fig. 2d). From this perspective, osteoblasts overlie new mineral on the endosteal surface (Fig. 2d). Osteocytes are also visible in the histological view, occupying sites within the cortical bone (Fig. 2d, e, white arrow). The histological description taken together with the dorsal view obtained by stereomicroscopy more rigorously defines the 3D microanatomy. The section pictured in Figure 2 shows marrow spaces ∼100 μm from the periosteal surface of the bone to their center. Due to light scattering, the two-photon microscope cannot image completely through the calvarial bone, however, marrow spaces are routinely reached. The white bar in Figure 2d indicates the maximum depth (139 μm in this case) of the two-photon stack shown in Figure 3a.
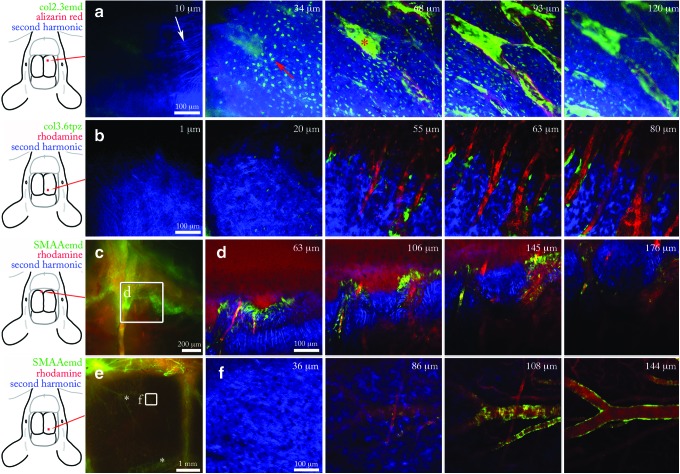
FIG. 3.
Two-photon live animal imaging of skeletal reporters in pristine bone. (a) Two-photon z-stack through mouse calvarial bone carrying a Col2.3emd reporter (green) and Alizarin mineral label (red). Red square on leftmost diagram indicates the x-y imaging location. Ten micrometer slice shows collagen fibers, white arrow. Thirty-four micrometer slice contains osteocytes, red arrow. (b) Parietal bone carrying a Col3.6topaz reporter (green) and rhodamine-labeled dextran in the vasculature (red). Osteoblasts localize in the area between the vasculature and the endosteal surface of bone. (c) Stereomicroscopy (leftmost panel) and two-photon z-stack of suture line containing SMAAemd osteoprogenitors (green) and rhodamine- labeled blood vessels (red). (d) z-Stack of SMAAemd osteoprogenitors shown in inset of (c). (e) Stereomicroscopy (leftmost panel) and two-photon z-stack of large blood vessels (*) in the parietal bone labeled with SMAAemd. Asterisks in (e) indicate SMAA+ blood vessels. (f) Smooth muscle cells labeled with SMAAemd wrap around rhodamine-labeled blood vessels. Color images available online at www.liebertpub.com/tec
Last, the two-photon z-stack previously taken of the parietal bone from the same animal shows a woven pattern of collagen fibers at the surface of the calvarium, visible by its second harmonic signal25–28 (Fig. 3a, 10 μm). As the scan moves into the calvarium, many dendritic osteocytes were visible occupying lacunae in the cortical bone (Fig. 3a, 34 μm). Moving into the marrow space (Fig. 3, 64 and 93 μm), Col2.3emd osteoblasts are seen at high density lining the endosteal surface labeled in red. At 120 μm, the sharpness of the image begins to degrade due to scattering in the tissue (Fig. 3, 120 μm). The description of the calvarial microanatomy with stereomicroscopy, histology, and two-photon microscopy provides a robust framework for the interpretation of two-photon data when viewed alone.
Two-photon imaging of skeletal lineage reporters
We used the reporters SMAA43 (osteoprogenitors), Col3.644 (early osteoblasts), and Col2.342,44 (mature osteoblasts and osteocytes) (Fig. 1a), to describe their baseline expression in the cellular microenvironment before use in later experiments. The starting tissue is rarely characterized before a regeneration study, yet this is an important step in understanding where responsible cells may originate. Col3.6 osteoblasts are present in the marrow spaces of the parietal bone and inside the suture lines. Blood vessels residing in the marrow space were visualized with a rhodamine injection (Fig. 3b). The SMAA transgene marks vascular lining cells, myofibroblasts, and a population of osteoprogenitor cells.43 We observed SMAAemd cells in the suture lines (Fig. 3c, d) and in the smooth muscle lining of blood vessels (Fig. 3e, f). This result implies that in a repair scenario involving resident SMAA osteoprogenitors, these cells would have to migrate from the suture lines to the site of injury. Taken together, these data form a part of the host's cellular initial conditions preceding a bone defect in the calvarium.
The collagen content and organization contribute to the mechanical properties, mass transport, and cell motility of living tissue.45 Collagen is readily observed in the calvarium by its generation of a second harmonic.25–28 We observed distinct features of collagen organization in the calvarial suture microenvironment. On the upper surface of the calvarium, the periosteal membrane contained highly organized collagen fibers in a woven pattern (Fig. 4, red asterisk, 19 μm). Deeper down, inside the cortical bone (Fig. 4, b-bone, 63–109 μm), collagen within mineralized tissue produces a more uniform second harmonic signal than nonmineralized membranous collagen. Within the suture space, there are many strands of wavy collagen fibers. These strands, collectively known as Sharpey's fibers,46 terminate in bundles along the wall of the marrow space at a regular interval (Fig. 4, 109 μm). Second harmonic imaging also allowed stain-free visualization of collagen-based scaffolds (Supplementary Fig. S2).25 These results provide the template of collagen in the periosteal membrane, suture space, and cortical bone in the host calvarial microenvironment preceding injury.
FIG. 4.
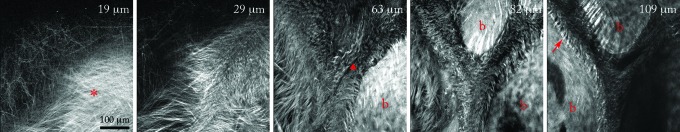
Collagen organization in the calvarial suture microenvironment. The second harmonic signal shows the organization of collagen fibers in the suture space and surrounding calvarial bone. Nineteen and 29 μm: the top layer of the calvarium shows woven collagen fibers of the periostem (red asterisk). Sixty-three micrometers: woven fibers give way to looser fibers inside the marrow space (red triangle). Cortical bone shows a dense second harmonic signal with the exception of bright streaking at insertion sites of collagen fibers from the marrow space into bone. Eighty-two micrometers: wavy fibers within the marrow space, surrounded by cortical bone (b). One hundred nine micrometers: bundles of collagen fibers, known as Sharpey's fibers, insert into the bone (red arrow). Fiber bundles (red arrow) appear at intervals along the marrow wall. Color images available online at www.liebertpub.com/tec
Tissue-engineered defect repair
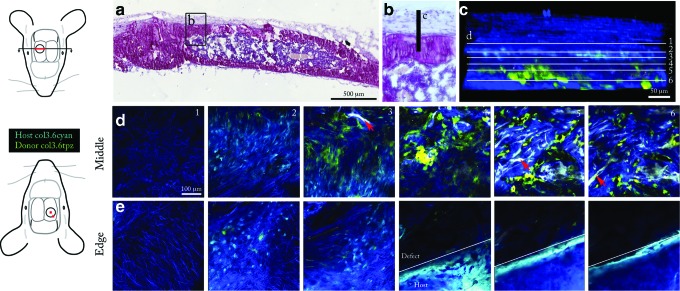
At 4 and 6 weeks postimplantation of a scaffold seeded with BMSCs, animals were observed with two-photon microscopy (Fig. 1b). Two locations were examined in each animal: an area in the central part of the implant and at the edge of the defect. At week 4, in the central part of the defect, a woven layer of collagen containing weakly expressing Col3.6cyan host cells with spindle-shaped morphology surrounded the implant on the top surface (Fig. 5d, panel 2). Host cell density was high in this layer above the defect. Relatively few donor cells were observed in this fibrous layer. Underneath the fibrous layer, scaffold and donor cells were visible (Fig. 5d, panels 4–6, and Supplementary Movie S1). Due to its spectrally broad autofluorescence, the scaffold appeared whitish in color, distinguishable from cells and the second harmonic signal (Fig. 5d, red arrows). A second harmonic signal similar to that observed in native bone was observed overlying the scaffold surface, suggestive of collagen in new mineral. The apposition rate of this layer on the scaffold surface was 0.69±0.48 μm/day (n=3, in one sample the implant was completely resorbed). Bright Col3.6topaz donor cells with osteoblastic cuboidal morphology populated the scaffold region, with some cells fully embedded in the new layer covering the scaffold surface. Donor cell density appeared moderate, with clusters of cells overlying the second harmonic layer indicative of new mineral. At the perimeter of the host bone 4 weeks after implantation, again a layer of host cells populated a fibrous layer above the scaffold and host bone. Similar to the central defect region, a few donor cells were visible in the fibrous layer (Fig. 5e). In contrast to more disparate clustering of donor cells in the implantation site, host cells formed a dense layer of bright Col3.6cyan osteoblasts on the surface of the host bone. Host cell density and cell–cell contact appeared to be greater on the surface of host bone than donor cells on the scaffold in the central defect region (Fig. 5d, e, panel 4 and below). This dense layer of host cells appeared to be depositing new mineral on the edge of host bone toward the central implant region (Supplementary Fig. S1). The MAR calculated from the thickness of the new mineral layer and the time since implantation was 1.88±0.13 μm/day (n=2, the edge of the original defect was not visible in the other samples). Some host cells were observed embedded in mineral behind the mineralization front. These results indicate that bone formation had occurred on the scaffold, predominantly by donor cells, and on the host bone, predominantly by host cells, at 4 weeks postimplantation.
FIG. 5.
Donor cells mineralize scaffold in vivo. (a) Hematoxylin-stained section of the implant obtained after the conclusion of imaging. Diagram at left shows the location of the cross section, highlighted in red, with respect to the circular defect. Inset (b) shows the area viewed by two-photon live animal microscopy (c). (c) Three-dimensional reconstruction of two-photon z-stack viewed side-on providing a histological orientation. White lines indicate the position of the six slices shown in (d). (d) Two-photon z-stack through the middle of the defect area at 4 weeks postimplantation. Diagram at left shows the location of the z-stack with respect to the defect (red square in middle of defect). Host cells are seen populating a layer of collagen fibers above the implant area (slice 2). The scaffold is visible due to autofluorescence (slice 3, 5, and 6, red arrow) and has a dense second harmonic signal on the surface, shown in blue. Donor cells populate the implant region (slice 4, 5, and 6). Z depths are 54, 109, 134, 156, 178, and 197 μm, respectively (panels 1–6). (e) Two-photon z-stack through the edge of the defect area at 4 weeks postimplantation. A layer of host cells covers the top surface (slice 2nd from left). Deeper into the stack, host osteoblasts form a dense layer on the surface of host bone. Z depths are 71, 119 145, 195, 245, and 275 μm, respectively. All two-photon images share the scale bar shown in the leftmost tile of (d). Color images available online at www.liebertpub.com/tec
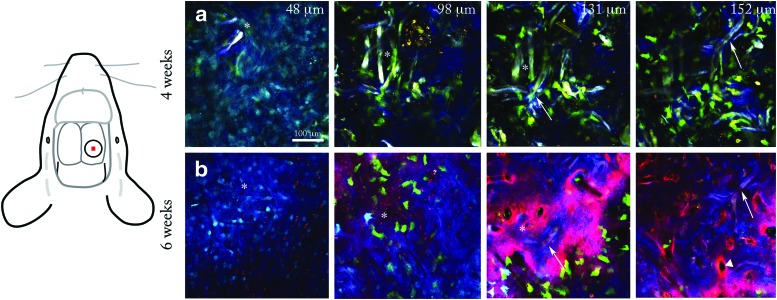
Using the pattern of the scaffold network as a guide, we returned to the same central defect location 2 weeks later (Fig. 6, compare two rightmost columns, arrows indicate identical features). At week 6, additional mineral was deposited in the implant center indicated by a large increase in the second harmonic layer, indicative of collagen within mineral (Fig. 6, compare two rightmost columns). Furthermore, the scaffold near the top surface was removed (Fig. 6, compare two leftmost columns, asterisks indicate identical locations), while deeper down, the scaffold was retained in mineral. Vertical canals were visible within the second harmonic surface labeled with Alizarin complexone (Fig. 6b, rightmost panel, white triangle). The colocalization of red mineral label to the second harmonic layer covering the scaffold confirms this tissue is indeed mineralized (Fig. 6b, two rightmost panels). From 4 to 6 weeks, donor cell density appeared reduced in two animals, while it remained similar in one animal. Bright Col3.6cyan host cells were observed on the mineral surface in the animal that retained moderate cell density. Taken together, these results indicate an increase in mineral deposition from 4 to 6 weeks and provide evidence of site-specific scaffold removal.
FIG. 6.
Comparison of scaffold and mineral regions at 4 and 6 weeks postimplantation. Two z-stacks taken at an identical location in the middle of the defect at 4 (a) and 6 weeks (b). Diagram at left shows the location of the stack with respect to the defect (red square). The scaffold on the top surface was removed between 4 and 6 weeks postimplantation (white asterisks indicate identical locations at 4 and 6 weeks). Deeper into the implant, the scaffold was retained (white arrows, slice 131 and 152 μm. A red mineralization label at 6 weeks (b) shows vertical channels in the matrix (white triangle, rightmost panel). Color images available online at www.liebertpub.com/tec
After imaging at week 6, calvaria from both the imaged and control group (no imaging or surgery) were harvested, X-rayed, and cut into thin sections. By examining the X-ray micrographs, better integration of the implant with host bone was observed in the control group than the test group (Fig. 7a–c). However, by histomorphometry, the area fraction of bone that formed in the defect was similar between the two groups (Figs. 7d–f). The resultant bone formed a core-shell architecture. Hematopoietic cells were observed in the core region (Supplementary Fig. S3), suggesting the marrow space was functional. Col3.6 osteoblasts were visible lining the inside and outside of the cortical shell (Fig. 7d, e). These data indicate that imaging surgery to expose the calvarium has a negative effect on host integration, but did not prohibit bone formation.
FIG. 7.
Effect of imaging surgery on bone formation. (a) X-ray of calvarium in the imaged group and (b) control group. (c) Radiopacity as a function of distance from the edge of the defect for the control and imaged group (n=3). (d) Histological section of defect from the imaged group and (e) control group. Donor cells (green), host cells (blue), and mineral label (red). (f) The bone area fraction in the defect region. Horizontal lines indicate the mean value for each group. The plus sign represents the sample that did not form bone in the time preceding imaging and was therefore excluded from the calculation of the mean. Color images available online at www.liebertpub.com/tec
Discussion
Two-photon microscopy combined with transgenic reporter animals has enabled researchers to observe, in vivo, the spatial and temporal dynamics of cell turnover during bone homeostasis and regeneration,21 and single transplanted hematopoietic progenitor cells as they engraft in the bone marrow.19,20 We have established a live animal imaging model of tissue-engineered bone using two-photon microscopy and transgenic reporter animals. Using this approach, we examined host cells, donor cells, scaffold, and collagen matrix in vivo during a tissue-engineered repair. Following the implantation of a collagen–hydroxyapatite scaffold seeded with bone marrow cells, we observed the in vivo outcome 4 and 6 weeks after implantation. New bone had formed on the surface of the scaffold primarily by donor-derived osteoblasts carrying the Col3.6topaz reporter at 4 weeks postimplantation (Fig. 5d, panel 5). This indicates that a sufficient number of donor cells survived transplantation and went on to form a major contribution to osteogenesis in the repair site. This finding recapitulates Yu et al., who found transplanted Col3.6cyan calvarial osteoprogenitor cells survived and formed new bone in a calvarial defect filled with a collagen–hydroxyapatite scaffold.35 Additionally, Cowan et al., using X-Y chromosome detection, found donor cells contributed 84%–99% of new bone at 8 weeks following implantation.32
An important question in bone tissue engineering is how does the host contribute to repair? In the present study, an outer layer of host cells with fibroblastic morphology encapsulated the implant in collagen fibers. These cells were weakly expressing the Col3.6cyan reporter and may later contribute host osteoblasts to the implant. Host Col3.6cyan cells were found on the mineral layer surrounding the scaffold at both week 4 and 6, although at a much lower incidence than donor Col3.6topaz cells. Park et al., using live animal two-photon microscopy, found that MX1 osteoprogenitor cells rapidly filled a calvarial microfracture (<1 mm diameter) and differentiated into osteoblasts at the bone edge.21 In the present study, fibroblastic host cells stemmed from the periosteal surface of the host calvarium, seen by histological examination (data not shown). The periosteum is a well-known contributor to fracture repair of long bones.47 Due to orientation of the imaging plane, two-photon microscopy enabled unprecedented observation of the new periosteal layer formed by the host above the cell–scaffold construct, likely by a similar osteoprogenitor population found by Park et al.
An increase in mineral deposition indicated by the second harmonic signal was observed between 4 and 6 weeks postimplantation. Additionally, it appeared that the scaffold near the top surface was resorbed (Fig. 6, 48 and 98 μm). This may be due to cell-mediated proteolytic degradation of type I collagen fibers in the scaffold by matrix metalloproteinases, namely, MMP-1.48,49 By contrast, the scaffold deeper inside the implant was embedded in mineral and persisted for the duration of the experiment (Fig. 6, 131 and 152 μm). The mineral and cellular organization in the scaffold at week 6 is likely an intermediate stage before the bone is more fully remodeled. Thus, the embedded scaffold would only be removed after substantial remodeling of tissue-engineered bone. Yu et al. found that the scaffold structure influences the structure of newly formed bone.35 We found the matrix being directly deposited on the scaffold structure until the pores were filled. Vertical channels were also noted in the new cortical layer produced at 6 weeks. These channels presumably link blood vessels from outside the implant to the implant core. Live animal microscopy enabled high-resolution examination of implant microenvironments at two time points, showing new mineral apposition and scaffold remodeling. This approach could provide new insights on how other scaffold materials are degraded and how mineral is deposited on scaffold surfaces.
Differences in cell organization and density were observed between host cells on the defect edge and donor cells on the mineral surface surrounding the scaffold. At the edge of the host bone, Col3.6cyan host osteoblasts formed a dense layer of cells that formed bone inward toward the implant. In contrast, donor cell density on the scaffold surface appeared more moderate with disparate clusters of cells. Additionally, the MAR appeared to be greater at the host edge than in the scaffold region. More robust measurement of mineral apposition is needed, however, this would suggest that tissue-engineered bone formation is lagging behind host osteogenesis.
In the imaged group, there was lower scaffold–host integration. This may be due to additional inflammatory signals introduced as a byproduct of the imaging surgery. However, the amount of bone in the defect area was similar (Fig. 7d–f), and bone continued to form beyond the 4-week time point as shown by the increase of the second harmonic signal in Figure 6 (two rightmost columns). This suggests that despite the imaging surgery, the processes essential to osteogenesis are preserved. Therefore, important questions that exist within the bounds of osteogenesis, regarding angiogenesis, scaffold degradation, cell signaling, proliferation and differentiation, may be addressed with this method. In combination with fluorescent reporters, any genetic entity of interest may be examined in vivo during bone formation in a cell-seeded construct. By enabling researchers to probe osteogenesis in vivo, two-photon microscopy combined with fluorescent reporters should improve our understanding of the mechanisms that drive bone healing in a cell–scaffold construct.
In the future, new techniques and improved optics may enable imaging noninvasively through the skin. Recently, Tang et al. have enabled significant improvements in the imaging depth of multiphoton microscopy by active compensation and reduction of light scattering in thick heterogeneous tissue.50 In their work, they imaged completely through a mouse skull to visualize fluorescent beads under the bottom surface. Although this method is currently limited to small fields of view (∼10 μm), implementing image reconstruction algorithms should bring in vivo imaging to new depths within an arbitrarily large field of view. Additionally, the use of longer wavelength excitation (near infrared), corresponding to less light scattering, should also extend the maximum depth of two-photon imaging systems. However, this also depends on the development of probes capable of far-red excitation. Thus, future examination of cells in bone and bone defects may be performed noninvasively, eliminating imaging surgery and associated effects.
In summary, we have evaluated tissue-engineered bone in vivo at a single-cell resolution and in situ in three dimensions at two time points in the same animals. Using skeletal lineage reporters, we have described the tissue microenvironment preceding a repair study and enabled the connection between stereomicroscopy, histology, and two-photon microscopy. In doing so, we have also described the collagen organization in calvarial bone and marrow space. Last, we have revealed a window into the dynamic progression of a tissue-engineered construct in vivo, previously unseen. In this view, we have made observations of donor cells, host cells, scaffold, and collagen organization in the repair site. Visualizing the cell and biomaterial interactions that govern the regeneration process in vivo should contribute to the development of tissue-engineering therapies that efficiently utilize donor cells and cooperate with the host response.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Xi Jiang, Ms. Li Chen, Dr. Mark Kronenberg, Dr. Kamal Khanna, and Dr. Peter Maye for their technical expertise and helpful discussions. This work was supported by a GAANN predoctoral fellowship from the Department of Education (P200A09315) and the NIH (1R21AR059962-01A1).
Disclosure Statement
No competing financial interests exist.
References
- 1.Atala A. Bauer S.B. Soker S. Yoo J.J. Retik A.B. Tissue-engineered autologous bladders for patients needing cystoplasty. Lancet. 2006;367:1241. doi: 10.1016/S0140-6736(06)68438-9. [DOI] [PubMed] [Google Scholar]
- 2.Macchiarini P., et al. Clinical transplantation of a tissue-engineered airway. Lancet. 2008;372:2023. doi: 10.1016/S0140-6736(08)61598-6. [DOI] [PubMed] [Google Scholar]
- 3.Ott H.C., et al. Regeneration and orthotopic transplantation of a bioartificial lung. Nat Med. 2010;16:927. doi: 10.1038/nm.2193. [DOI] [PubMed] [Google Scholar]
- 4.Vogel G. Mending the youngest hearts. Science. 2011;333:1088. doi: 10.1126/science.333.6046.1088. [DOI] [PubMed] [Google Scholar]
- 5.Petersen T.H., et al. Tissue-engineered lungs for in vivo implantation. Science. 2010;329:538. doi: 10.1126/science.1189345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffith L.G. Tissue engineering—current challenges and expanding opportunities. Science. 2002;295:1009. doi: 10.1126/science.1069210. [DOI] [PubMed] [Google Scholar]
- 7.Muschler G.F. Raut V.P. Patterson T.E. Wenke J.C. Hollinger J.O. The design and use of animal models for translational research in bone tissue engineering and regenerative medicine. Tissue Eng Part B Rev. 2010;16:123. doi: 10.1089/ten.TEB.2009.0658. [DOI] [PubMed] [Google Scholar]
- 8.Beckman M. Play-by-play imaging rewrites cells' rules. Science. 2003;300:76. doi: 10.1126/science.300.5616.76. [DOI] [PubMed] [Google Scholar]
- 9.Holtmaat A., et al. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat Protoc. 2009;4:1128. doi: 10.1038/nprot.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mostany R. Portera-Cailliau C. A Craniotomy surgery procedure for chronic brain imaging. J Vis Exp. 2008;12 doi: 10.3791/680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mostany R. Portera-Cailliau C. A method for 2-photon imaging of blood flow in the neocortex through a cranial window. J Vis Exp. 2008;12 doi: 10.3791/678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seung H.S. Neuroscience: towards functional connectomics. Nature. 2011;471:170. doi: 10.1038/471170a. [DOI] [PubMed] [Google Scholar]
- 13.Khanna K.M. McNamara J.T. Lefrancois L. In situ imaging of the endogenous CD8 T cell response to infection. Science. 2007;318:116. doi: 10.1126/science.1146291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Celli S. Albert M.L. Bousso P. Visualizing the innate and adaptive immune responses underlying allograft rejection by two-photon microscopy. Nat Med. 2011;17:744. doi: 10.1038/nm.2376. [DOI] [PubMed] [Google Scholar]
- 15.Hickman H.D. Bennink J.R. Yewdell J.W. Caught in the act: intravital multiphoton microscopy of host-pathogen interactions. Cell Host Microbe. 2009;5:13. doi: 10.1016/j.chom.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cahalan M.D. Parker I. Wei S.H. Miller M.J. Two-photon tissue imaging: seeing the immune system in a fresh light. Nat Rev Immunol. 2002;2:872. doi: 10.1038/nri935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cahalan M.D. Parker I. Imaging the choreography of lymphocyte trafficking and the immune response. Curr Opin Immunol. 2006;18:476. doi: 10.1016/j.coi.2006.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W., et al. Intravital 2-photon imaging of leukocyte trafficking in beating heart. J Clin Invest. 2012;122:2499. doi: 10.1172/JCI62970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo Celso C., et al. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature. 2009;457:92. doi: 10.1038/nature07434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo Celso C. Lin C.P. Scadden D.T. In vivo imaging of transplanted hematopoietic stem and progenitor cells in mouse calvarium bone marrow. Nat Protoc. 2011;6:1. doi: 10.1038/nprot.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park D., et al. Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell. 2012;10:259. doi: 10.1016/j.stem.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Göppert-Mayer M. Über elementarakte mit zwei quantensprüngen. Ann Phys. 1931;401:273. [Google Scholar]
- 23.Denk W. Strickler J.H. Webb W.W. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 24.Zinselmeyer B.H., et al. Vol. 461. San Deigo, CA: Elsevier; 2009. Methods in Enzymology; pp. 349–378. [DOI] [PubMed] [Google Scholar]
- 25.Williams R.M. Zipfel W.R. Webb W.W. Interpreting second-harmonic generation images of collagen I fibrils. Biophys J. 2005;88:1377. doi: 10.1529/biophysj.104.047308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown E., et al. Dynamic imaging of collagen and its modulation in tumors in vivo using second-harmonic generation. Nat Med. 2003;9:796. doi: 10.1038/nm879. [DOI] [PubMed] [Google Scholar]
- 27.Prasad P.N. Williams D.J. New York: Wiley; 1991. Introduction to Nonlinear Optical Effects in Molecules and Polymers. [Google Scholar]
- 28.Cox G., et al. 3-Dimensional imaging of collagen using second harmonic generation. J Struct Biol. 2003;141:53. doi: 10.1016/s1047-8477(02)00576-2. [DOI] [PubMed] [Google Scholar]
- 29.Tsigkou O., et al. Engineered vascularized bone grafts. Proc Natl Acad Sci U S A. 2010;107:3311. doi: 10.1073/pnas.0905445107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martini J., et al. Two-photon laser scanning microscopy on native cartilage and collagen-membranes for tissue-engineering. In Proceedings of SPIE, Multiphoton Microscopy in the Biomedical Sciences; San Jose, CA. 2006. p. 60891N. [Google Scholar]
- 31.Aghaloo T., et al. The effect of NELL1 and bone morphogenetic protein-2 on calvarial bone regeneration. J Oral Maxillofac Surg. 2010;68:300. doi: 10.1016/j.joms.2009.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowan C.M., et al. Adipose-derived adult stromal cells heal critical-size mouse calvarial defects. Nat Biotechnol. 2004;22:560. doi: 10.1038/nbt958. [DOI] [PubMed] [Google Scholar]
- 33.Xu X. Yang Z. Liu Q. Wang Y. In vivo fluorescence imaging of muscle cell regeneration by transplanted EGFP-labeled myoblasts. Mol Ther. 2010;18:835. doi: 10.1038/mt.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroeder T. Imaging stem-cell-driven regeneration in mammals. Nature. 2008;453:345. doi: 10.1038/nature07043. [DOI] [PubMed] [Google Scholar]
- 35.Yu X., et al. Controlling the structural organization of regenerated bone by tailoring tissue engineering scaffold architecture. J Mater Chem. 2012;22:9721. [Google Scholar]
- 36.Yin D., et al. Determination of the fate and contribution of ex vivo expanded human bone marrow stem and progenitor cells for bone formation by 2.3ColGFP. Mol Ther. 2009;17:1967. doi: 10.1038/mt.2009.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu X., et al. The effect of fresh bone marrow cells on reconstruction of mouse calvarial defect combined with calvarial osteoprogenitor cells, collagen–apatite scaffold. J Tissue Eng Regen Med. 2012 doi: 10.1002/term.1490. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmid B. Schindelin J. Cardona A. Longair M. Heisenberg M. Software A high-level 3D visualization API for Java and ImageJ. 2010. http://3dviewer.neurofly.de. http://3dviewer.neurofly.de [DOI] [PMC free article] [PubMed]
- 39.Schindelin J., et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Advanced Weka Segmentation. http://fiji.sc/wiki/index.php/Advanced_Weka_Segmentation http://fiji.sc/wiki/index.php/Advanced_Weka_Segmentation
- 41.Hildebrand T. Rüegsegger P. A new method for the model-independent assessment of thickness in three-dimensional images. J Microsc. 1997;185:67. [Google Scholar]
- 42.Kalajzic Z., et al. Directing the expression of a green fluorescent protein transgene in differentiated osteoblasts: comparison between rat type I collagen and rat osteocalcin promoters. Bone. 2002;31:654. doi: 10.1016/s8756-3282(02)00912-2. [DOI] [PubMed] [Google Scholar]
- 43.Kalajzic Z., et al. Use of an alpha-smooth muscle actin GFP reporter to identify an osteoprogenitor population. Bone. 2008;43:501. doi: 10.1016/j.bone.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalajzic I., et al. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone Miner Res. 2002;17:15. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- 45.Blitterswijk C.A. van Thomsen P. London, UK: Academic Press; 2008. Tissue Engineering. [Google Scholar]
- 46.Aaron J.E. Periosteal Sharpey's fibers: a novel bone matrix regulatory system? Front Endocrinol. 2012;3:98. doi: 10.3389/fendo.2012.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bruder S.P. Fink D.J. Caplan A.I. Mesenchymal stem cells in bone development, bone repair, and skeletal regenaration therapy. J Cell Biochem. 1994;56:283. doi: 10.1002/jcb.240560809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pilcher B.K., et al. The activity of collagenase-1 is required for keratinocyte migration on a type I collagen matrix. J Cell Biol. 1997;137:1445. doi: 10.1083/jcb.137.6.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Page-McCaw A. Ewald A.J. Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8:221. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang J. Germain R.N. Cui M. Superpenetration optical microscopy by iterative multiphoton adaptive compensation technique. Proc Natl Acad Sci U S A. 2012;109:8434. doi: 10.1073/pnas.1119590109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.