Abstract
In the title compound, C25H21ClN2O2Se, the selenadiazole ring is almost planar [maximum deviation = 0.004 (2) Å], and the adjacent benzene ring is twisted by 50.6 (1)° with respect to this ring.
Related literature
For general background to selenadiazol derivatives, see: Khanna (2005 ▶). For related structures, see: Marx et al. (2008 ▶); Muthukumaran et al. (2011 ▶).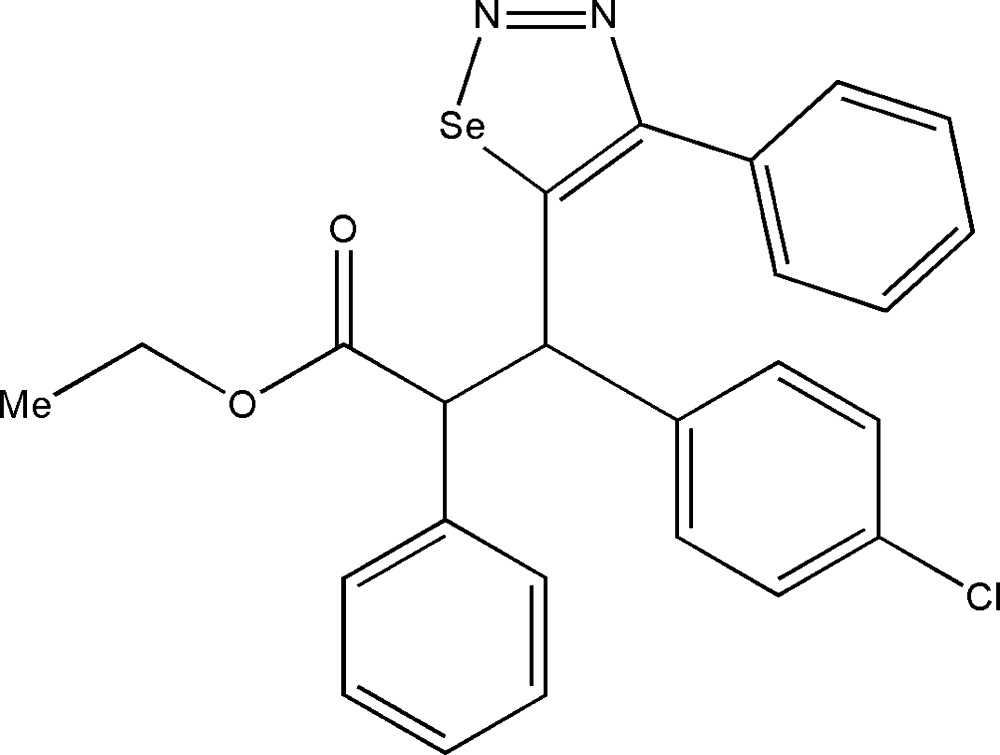
Experimental
Crystal data
C25H21ClN2O2Se
M r = 495.85
Monoclinic,

a = 12.1337 (3) Å
b = 12.2267 (3) Å
c = 16.4423 (4) Å
β = 107.744 (1)°
V = 2323.26 (10) Å3
Z = 4
Mo Kα radiation
μ = 1.76 mm−1
T = 293 K
0.25 × 0.20 × 0.18 mm
Data collection
Bruker SMART APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2008 ▶) T min = 0.663, T max = 0.729
22323 measured reflections
5764 independent reflections
3745 reflections with I > 2σ(I)
R int = 0.035
Refinement
R[F 2 > 2σ(F 2)] = 0.037
wR(F 2) = 0.107
S = 1.02
5764 reflections
280 parameters
H-atom parameters constrained
Δρmax = 0.42 e Å−3
Δρmin = −0.27 e Å−3
Data collection: APEX2 (Bruker, 2008 ▶); cell refinement: SAINT (Bruker, 2008 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▶); software used to prepare material for publication: SHELXL97 and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813017790/ng5333sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813017790/ng5333Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813017790/ng5333Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
PS thanks the UGC, New Delhi, for financial support in the form of a Research Fellowship in Science for Meritorious Students. The authors thank the TBI Consultancy, University of Madras, India, for the data collection.
supplementary crystallographic information
Comment
Selenadiazoles, having one selenium and two nitrogen atoms in a five membered ring, are the important class of organoselenium compounds utilized in the synthesis of semiconductor nanoparticles (Khanna, 2005). The crystal structure of the title compound is carried out to elucidate the conformational status of the molecule.
The ORTEP plot of the molecule is shown in Fig.1. The selenadiazol ring is planar[with maximum deviation for the atom N2 is -0.004 (2) Å]. The attached phenyl ring is twisted away at an angle of 50.6 (1)° with respect to selenadiazol ring. The bond lengths [Se1—N2] 1.874 (2) Å, [Se1—C8] 1.838 (2) Å & [Cl1—C13] 1.736 (2)° are comparable with the values reported in the literature (Marx et al. 2008; Muthukumaran et al. 2011). The bond C9—C16 is slightly lengthened due to streic interaction between the phenyl and chlorophenyl rings.
The dihedral angle between the selenadiazol and chlorophenyl ring is 74.3 (1)°. The propanoate group assumes an extended conformation which can be seen from the torsion angle (C16—C23—O2—C24) value of -177.8 (2)°.
Experimental
A mixture of ethyl 3-(4-chlorophenyl)-5-oxo-2,5-diphenylpentanoate (1 mM), semicarbazide hydrochloride(2 mM) and sodium acetate (3 mM) in ethanol (10 ml) was refluxed for 4 hrs. After completion of the reaction as monitored by TLC, the mixture was poured into ice cold water and the resulting semicarbazone was filtered off. Then, a mixture of semicarbazone (1 mM) and SeO2 (2 mM) in tetrahydrofuran (10 ml) were refluxed on a water bath for 1hr. The selenium deposited on cooling was removed by filtration, and the filtrate was poured into crushed ice, extracted with dichloromethane, and purified by column chromatography using silica gel (60–120 mesh) with 97:3 petroleum ether: ethyl acetate as eluent to give ethyl-3-(4-chlorophenyl)-2-phenyl-3- (4-phenyl-1,2, 3-selenadiazol-5-yl)propanoate.
Refinement
H atoms were positioned geometrically (C—H = 0.93–0.98 Å) and allowed to ride on their parent atoms, with Uiso(H) = 1.5Ueq(C) for methyl H and 1.2Ueq(C) for other H atoms.
Figures
Fig. 1.
The molecular structure of the title compound with the displacement ellipsoids drawn at 30% probability level.
Crystal data
| C25H21ClN2O2Se | F(000) = 1008 |
| Mr = 495.85 | Dx = 1.418 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 5764 reflections |
| a = 12.1337 (3) Å | θ = 1.8–28.4° |
| b = 12.2267 (3) Å | µ = 1.76 mm−1 |
| c = 16.4423 (4) Å | T = 293 K |
| β = 107.744 (1)° | Block, yellow |
| V = 2323.26 (10) Å3 | 0.25 × 0.20 × 0.18 mm |
| Z = 4 |
Data collection
| Bruker SMART APEXII CCD diffractometer | 5764 independent reflections |
| Radiation source: fine-focus sealed tube | 3745 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.035 |
| ω and φ scans | θmax = 28.4°, θmin = 1.8° |
| Absorption correction: multi-scan (SADABS; Bruker, 2008) | h = −12→16 |
| Tmin = 0.663, Tmax = 0.729 | k = −16→15 |
| 22323 measured reflections | l = −21→19 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.037 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.107 | H-atom parameters constrained |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.0471P)2 + 0.6047P] where P = (Fo2 + 2Fc2)/3 |
| 5764 reflections | (Δ/σ)max = 0.002 |
| 280 parameters | Δρmax = 0.42 e Å−3 |
| 0 restraints | Δρmin = −0.27 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.7760 (2) | 0.5374 (2) | 0.74986 (16) | 0.0633 (6) | |
| H1 | 0.7656 | 0.5310 | 0.6916 | 0.076* | |
| C2 | 0.7134 (3) | 0.6137 (2) | 0.7793 (2) | 0.0836 (9) | |
| H2 | 0.6610 | 0.6588 | 0.7407 | 0.100* | |
| C3 | 0.7278 (3) | 0.6234 (3) | 0.8652 (3) | 0.0954 (10) | |
| H3 | 0.6847 | 0.6744 | 0.8846 | 0.114* | |
| C4 | 0.8054 (3) | 0.5580 (3) | 0.9219 (2) | 0.0908 (10) | |
| H4 | 0.8151 | 0.5647 | 0.9800 | 0.109* | |
| C5 | 0.8696 (2) | 0.4818 (2) | 0.89387 (17) | 0.0706 (7) | |
| H5 | 0.9230 | 0.4381 | 0.9331 | 0.085* | |
| C6 | 0.85467 (19) | 0.47046 (18) | 0.80734 (14) | 0.0536 (5) | |
| C7 | 0.92554 (18) | 0.38995 (18) | 0.77867 (13) | 0.0502 (5) | |
| C8 | 0.88943 (17) | 0.31106 (18) | 0.71769 (13) | 0.0469 (5) | |
| C9 | 0.76586 (16) | 0.28344 (17) | 0.66740 (12) | 0.0428 (4) | |
| H9 | 0.7191 | 0.3492 | 0.6659 | 0.051* | |
| C10 | 0.72046 (16) | 0.19519 (17) | 0.71371 (12) | 0.0426 (4) | |
| C11 | 0.64815 (19) | 0.2233 (2) | 0.76108 (15) | 0.0549 (5) | |
| H11 | 0.6269 | 0.2960 | 0.7634 | 0.066* | |
| C12 | 0.6070 (2) | 0.1451 (2) | 0.80495 (15) | 0.0656 (7) | |
| H12 | 0.5585 | 0.1648 | 0.8367 | 0.079* | |
| C13 | 0.6383 (2) | 0.0383 (2) | 0.80109 (15) | 0.0631 (7) | |
| C14 | 0.7116 (2) | 0.0082 (2) | 0.75615 (15) | 0.0622 (6) | |
| H14 | 0.7338 | −0.0644 | 0.7551 | 0.075* | |
| C15 | 0.7521 (2) | 0.08659 (19) | 0.71257 (14) | 0.0538 (5) | |
| H15 | 0.8016 | 0.0663 | 0.6818 | 0.065* | |
| C16 | 0.75310 (17) | 0.25174 (17) | 0.57420 (13) | 0.0456 (5) | |
| H16 | 0.7931 | 0.1820 | 0.5747 | 0.055* | |
| C17 | 0.62689 (19) | 0.23681 (19) | 0.52183 (13) | 0.0497 (5) | |
| C18 | 0.5471 (2) | 0.3186 (2) | 0.51442 (14) | 0.0640 (6) | |
| H18 | 0.5699 | 0.3853 | 0.5415 | 0.077* | |
| C19 | 0.4325 (2) | 0.3023 (3) | 0.46674 (16) | 0.0834 (9) | |
| H19 | 0.3785 | 0.3579 | 0.4619 | 0.100* | |
| C20 | 0.3993 (3) | 0.2040 (4) | 0.4268 (2) | 0.0985 (12) | |
| H20 | 0.3222 | 0.1923 | 0.3958 | 0.118* | |
| C21 | 0.4772 (3) | 0.1247 (3) | 0.4322 (2) | 0.1093 (13) | |
| H21 | 0.4542 | 0.0589 | 0.4037 | 0.131* | |
| C22 | 0.5919 (3) | 0.1399 (2) | 0.47981 (18) | 0.0818 (8) | |
| H22 | 0.6454 | 0.0842 | 0.4833 | 0.098* | |
| C23 | 0.80904 (19) | 0.3363 (2) | 0.53277 (14) | 0.0533 (5) | |
| C24 | 0.9294 (3) | 0.3559 (3) | 0.4427 (2) | 0.0910 (9) | |
| H24A | 0.9033 | 0.3406 | 0.3820 | 0.109* | |
| H24B | 0.9149 | 0.4325 | 0.4509 | 0.109* | |
| C25 | 1.0522 (3) | 0.3337 (3) | 0.4767 (3) | 0.1152 (13) | |
| H25A | 1.0935 | 0.3782 | 0.4477 | 0.173* | |
| H25B | 1.0663 | 0.2579 | 0.4683 | 0.173* | |
| H25C | 1.0781 | 0.3502 | 0.5366 | 0.173* | |
| N1 | 1.04418 (17) | 0.39168 (19) | 0.81995 (13) | 0.0663 (5) | |
| N2 | 1.10574 (18) | 0.3221 (2) | 0.79702 (15) | 0.0766 (6) | |
| O1 | 0.80184 (17) | 0.43304 (16) | 0.54165 (12) | 0.0777 (5) | |
| O2 | 0.86582 (16) | 0.28857 (15) | 0.48558 (11) | 0.0679 (5) | |
| Cl1 | 0.58491 (9) | −0.06117 (8) | 0.85433 (6) | 0.1057 (3) | |
| Se1 | 1.01461 (2) | 0.23132 (3) | 0.710678 (17) | 0.06928 (12) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0524 (13) | 0.0566 (14) | 0.0712 (15) | −0.0052 (11) | 0.0043 (11) | −0.0039 (12) |
| C2 | 0.0652 (17) | 0.0621 (17) | 0.109 (2) | 0.0021 (14) | 0.0058 (16) | −0.0044 (16) |
| C3 | 0.090 (2) | 0.079 (2) | 0.120 (3) | 0.0026 (18) | 0.036 (2) | −0.025 (2) |
| C4 | 0.119 (3) | 0.079 (2) | 0.080 (2) | −0.015 (2) | 0.039 (2) | −0.0217 (17) |
| C5 | 0.0822 (19) | 0.0585 (16) | 0.0642 (16) | −0.0078 (13) | 0.0121 (13) | −0.0058 (12) |
| C6 | 0.0494 (12) | 0.0467 (12) | 0.0576 (13) | −0.0146 (10) | 0.0057 (10) | −0.0040 (10) |
| C7 | 0.0385 (11) | 0.0530 (13) | 0.0515 (12) | −0.0106 (9) | 0.0025 (9) | 0.0027 (10) |
| C8 | 0.0350 (10) | 0.0535 (12) | 0.0485 (11) | −0.0031 (9) | 0.0071 (8) | 0.0028 (9) |
| C9 | 0.0331 (9) | 0.0476 (12) | 0.0437 (11) | −0.0031 (8) | 0.0056 (8) | −0.0046 (8) |
| C10 | 0.0336 (9) | 0.0515 (12) | 0.0391 (10) | −0.0046 (8) | 0.0058 (8) | −0.0060 (8) |
| C11 | 0.0436 (11) | 0.0644 (14) | 0.0559 (13) | 0.0018 (10) | 0.0142 (10) | −0.0053 (11) |
| C12 | 0.0503 (13) | 0.093 (2) | 0.0591 (14) | −0.0091 (13) | 0.0247 (11) | −0.0038 (13) |
| C13 | 0.0588 (14) | 0.0749 (18) | 0.0536 (13) | −0.0249 (13) | 0.0141 (11) | 0.0025 (12) |
| C14 | 0.0702 (16) | 0.0545 (14) | 0.0609 (14) | −0.0117 (12) | 0.0185 (12) | −0.0019 (11) |
| C15 | 0.0561 (13) | 0.0543 (14) | 0.0546 (13) | −0.0043 (10) | 0.0222 (10) | −0.0054 (10) |
| C16 | 0.0383 (10) | 0.0516 (13) | 0.0451 (11) | −0.0037 (9) | 0.0098 (8) | −0.0024 (9) |
| C17 | 0.0447 (11) | 0.0649 (14) | 0.0365 (10) | −0.0117 (10) | 0.0082 (8) | −0.0027 (9) |
| C18 | 0.0468 (13) | 0.0940 (19) | 0.0471 (13) | 0.0022 (13) | 0.0082 (10) | −0.0126 (12) |
| C19 | 0.0429 (13) | 0.148 (3) | 0.0523 (14) | 0.0061 (16) | 0.0045 (11) | −0.0008 (16) |
| C20 | 0.0558 (18) | 0.154 (4) | 0.0665 (18) | −0.038 (2) | −0.0093 (14) | 0.011 (2) |
| C21 | 0.097 (3) | 0.095 (3) | 0.101 (2) | −0.043 (2) | −0.024 (2) | −0.007 (2) |
| C22 | 0.0739 (18) | 0.0680 (18) | 0.0809 (18) | −0.0149 (14) | −0.0103 (14) | −0.0079 (14) |
| C23 | 0.0448 (12) | 0.0626 (16) | 0.0478 (12) | −0.0083 (10) | 0.0073 (9) | 0.0007 (10) |
| C24 | 0.085 (2) | 0.114 (3) | 0.086 (2) | −0.0062 (19) | 0.0439 (17) | 0.0262 (18) |
| C25 | 0.087 (3) | 0.094 (3) | 0.179 (4) | −0.016 (2) | 0.062 (3) | 0.019 (2) |
| N1 | 0.0416 (11) | 0.0765 (14) | 0.0671 (13) | −0.0124 (10) | −0.0039 (9) | −0.0003 (10) |
| N2 | 0.0349 (10) | 0.0998 (18) | 0.0827 (15) | −0.0066 (11) | −0.0002 (10) | −0.0018 (13) |
| O1 | 0.0910 (14) | 0.0613 (12) | 0.0898 (13) | −0.0124 (10) | 0.0409 (11) | 0.0029 (9) |
| O2 | 0.0688 (11) | 0.0807 (12) | 0.0641 (10) | −0.0032 (9) | 0.0352 (9) | 0.0054 (9) |
| Cl1 | 0.1182 (7) | 0.1091 (7) | 0.1002 (6) | −0.0457 (5) | 0.0485 (5) | 0.0149 (5) |
| Se1 | 0.03892 (14) | 0.0867 (2) | 0.0777 (2) | 0.00811 (12) | 0.01107 (11) | −0.00725 (13) |
Geometric parameters (Å, º)
| C1—C2 | 1.380 (4) | C14—H14 | 0.9300 |
| C1—C6 | 1.387 (3) | C15—H15 | 0.9300 |
| C1—H1 | 0.9300 | C16—C23 | 1.509 (3) |
| C2—C3 | 1.374 (4) | C16—C17 | 1.522 (3) |
| C2—H2 | 0.9300 | C16—H16 | 0.9800 |
| C3—C4 | 1.364 (5) | C17—C18 | 1.372 (3) |
| C3—H3 | 0.9300 | C17—C22 | 1.372 (3) |
| C4—C5 | 1.381 (4) | C18—C19 | 1.386 (3) |
| C4—H4 | 0.9300 | C18—H18 | 0.9300 |
| C5—C6 | 1.385 (3) | C19—C20 | 1.371 (5) |
| C5—H5 | 0.9300 | C19—H19 | 0.9300 |
| C6—C7 | 1.476 (3) | C20—C21 | 1.338 (5) |
| C7—C8 | 1.364 (3) | C20—H20 | 0.9300 |
| C7—N1 | 1.392 (3) | C21—C22 | 1.386 (4) |
| C8—C9 | 1.513 (3) | C21—H21 | 0.9300 |
| C8—Se1 | 1.838 (2) | C22—H22 | 0.9300 |
| C9—C10 | 1.518 (3) | C23—O1 | 1.198 (3) |
| C9—C16 | 1.542 (3) | C23—O2 | 1.320 (3) |
| C9—H9 | 0.9800 | C24—C25 | 1.448 (5) |
| C10—C11 | 1.382 (3) | C24—O2 | 1.449 (3) |
| C10—C15 | 1.384 (3) | C24—H24A | 0.9700 |
| C11—C12 | 1.379 (3) | C24—H24B | 0.9700 |
| C11—H11 | 0.9300 | C25—H25A | 0.9600 |
| C12—C13 | 1.367 (4) | C25—H25B | 0.9600 |
| C12—H12 | 0.9300 | C25—H25C | 0.9600 |
| C13—C14 | 1.368 (3) | N1—N2 | 1.263 (3) |
| C13—Cl1 | 1.736 (2) | N2—Se1 | 1.874 (2) |
| C14—C15 | 1.374 (3) | ||
| C2—C1—C6 | 119.8 (3) | C14—C15—H15 | 119.4 |
| C2—C1—H1 | 120.1 | C10—C15—H15 | 119.4 |
| C6—C1—H1 | 120.1 | C23—C16—C17 | 109.90 (17) |
| C3—C2—C1 | 120.5 (3) | C23—C16—C9 | 110.70 (17) |
| C3—C2—H2 | 119.7 | C17—C16—C9 | 111.81 (17) |
| C1—C2—H2 | 119.7 | C23—C16—H16 | 108.1 |
| C4—C3—C2 | 119.8 (3) | C17—C16—H16 | 108.1 |
| C4—C3—H3 | 120.1 | C9—C16—H16 | 108.1 |
| C2—C3—H3 | 120.1 | C18—C17—C22 | 118.9 (2) |
| C3—C4—C5 | 120.6 (3) | C18—C17—C16 | 121.6 (2) |
| C3—C4—H4 | 119.7 | C22—C17—C16 | 119.5 (2) |
| C5—C4—H4 | 119.7 | C17—C18—C19 | 120.3 (3) |
| C4—C5—C6 | 120.0 (3) | C17—C18—H18 | 119.8 |
| C4—C5—H5 | 120.0 | C19—C18—H18 | 119.8 |
| C6—C5—H5 | 120.0 | C20—C19—C18 | 119.7 (3) |
| C5—C6—C1 | 119.2 (2) | C20—C19—H19 | 120.2 |
| C5—C6—C7 | 119.2 (2) | C18—C19—H19 | 120.2 |
| C1—C6—C7 | 121.5 (2) | C21—C20—C19 | 120.3 (3) |
| C8—C7—N1 | 115.0 (2) | C21—C20—H20 | 119.8 |
| C8—C7—C6 | 128.22 (19) | C19—C20—H20 | 119.8 |
| N1—C7—C6 | 116.80 (19) | C20—C21—C22 | 120.6 (3) |
| C7—C8—C9 | 127.0 (2) | C20—C21—H21 | 119.7 |
| C7—C8—Se1 | 109.56 (15) | C22—C21—H21 | 119.7 |
| C9—C8—Se1 | 123.19 (16) | C17—C22—C21 | 120.2 (3) |
| C8—C9—C10 | 109.59 (16) | C17—C22—H22 | 119.9 |
| C8—C9—C16 | 112.45 (17) | C21—C22—H22 | 119.9 |
| C10—C9—C16 | 112.21 (16) | O1—C23—O2 | 125.4 (2) |
| C8—C9—H9 | 107.4 | O1—C23—C16 | 124.2 (2) |
| C10—C9—H9 | 107.4 | O2—C23—C16 | 110.4 (2) |
| C16—C9—H9 | 107.4 | C25—C24—O2 | 110.1 (3) |
| C11—C10—C15 | 118.2 (2) | C25—C24—H24A | 109.6 |
| C11—C10—C9 | 119.7 (2) | O2—C24—H24A | 109.6 |
| C15—C10—C9 | 122.06 (19) | C25—C24—H24B | 109.6 |
| C12—C11—C10 | 121.1 (2) | O2—C24—H24B | 109.6 |
| C12—C11—H11 | 119.5 | H24A—C24—H24B | 108.2 |
| C10—C11—H11 | 119.5 | C24—C25—H25A | 109.5 |
| C13—C12—C11 | 119.1 (2) | C24—C25—H25B | 109.5 |
| C13—C12—H12 | 120.4 | H25A—C25—H25B | 109.5 |
| C11—C12—H12 | 120.4 | C24—C25—H25C | 109.5 |
| C14—C13—C12 | 121.2 (2) | H25A—C25—H25C | 109.5 |
| C14—C13—Cl1 | 119.2 (2) | H25B—C25—H25C | 109.5 |
| C12—C13—Cl1 | 119.6 (2) | N2—N1—C7 | 117.5 (2) |
| C13—C14—C15 | 119.3 (2) | N1—N2—Se1 | 111.03 (15) |
| C13—C14—H14 | 120.4 | C23—O2—C24 | 119.0 (2) |
| C15—C14—H14 | 120.4 | C8—Se1—N2 | 86.93 (10) |
| C14—C15—C10 | 121.2 (2) | ||
| C6—C1—C2—C3 | −0.4 (4) | C11—C10—C15—C14 | 1.0 (3) |
| C1—C2—C3—C4 | 0.7 (5) | C9—C10—C15—C14 | 178.84 (19) |
| C2—C3—C4—C5 | −0.1 (5) | C8—C9—C16—C23 | −50.6 (2) |
| C3—C4—C5—C6 | −0.7 (5) | C10—C9—C16—C23 | −174.72 (17) |
| C4—C5—C6—C1 | 1.0 (4) | C8—C9—C16—C17 | −173.52 (18) |
| C4—C5—C6—C7 | 179.0 (2) | C10—C9—C16—C17 | 62.4 (2) |
| C2—C1—C6—C5 | −0.5 (4) | C23—C16—C17—C18 | −66.8 (3) |
| C2—C1—C6—C7 | −178.4 (2) | C9—C16—C17—C18 | 56.6 (3) |
| C5—C6—C7—C8 | 129.5 (3) | C23—C16—C17—C22 | 112.2 (3) |
| C1—C6—C7—C8 | −52.6 (3) | C9—C16—C17—C22 | −124.4 (2) |
| C5—C6—C7—N1 | −48.6 (3) | C22—C17—C18—C19 | 1.4 (4) |
| C1—C6—C7—N1 | 129.3 (2) | C16—C17—C18—C19 | −179.6 (2) |
| N1—C7—C8—C9 | 174.8 (2) | C17—C18—C19—C20 | −0.1 (4) |
| C6—C7—C8—C9 | −3.3 (4) | C18—C19—C20—C21 | −1.4 (5) |
| N1—C7—C8—Se1 | −0.1 (2) | C19—C20—C21—C22 | 1.6 (6) |
| C6—C7—C8—Se1 | −178.24 (18) | C18—C17—C22—C21 | −1.3 (4) |
| C7—C8—C9—C10 | −91.3 (3) | C16—C17—C22—C21 | 179.7 (3) |
| Se1—C8—C9—C10 | 83.0 (2) | C20—C21—C22—C17 | −0.2 (6) |
| C7—C8—C9—C16 | 143.2 (2) | C17—C16—C23—O1 | 82.5 (3) |
| Se1—C8—C9—C16 | −42.5 (2) | C9—C16—C23—O1 | −41.5 (3) |
| C8—C9—C10—C11 | 101.5 (2) | C17—C16—C23—O2 | −97.2 (2) |
| C16—C9—C10—C11 | −132.8 (2) | C9—C16—C23—O2 | 138.78 (19) |
| C8—C9—C10—C15 | −76.3 (2) | C8—C7—N1—N2 | 0.5 (3) |
| C16—C9—C10—C15 | 49.4 (2) | C6—C7—N1—N2 | 178.9 (2) |
| C15—C10—C11—C12 | −1.1 (3) | C7—N1—N2—Se1 | −0.7 (3) |
| C9—C10—C11—C12 | −178.98 (19) | O1—C23—O2—C24 | 2.6 (4) |
| C10—C11—C12—C13 | −0.1 (4) | C16—C23—O2—C24 | −177.8 (2) |
| C11—C12—C13—C14 | 1.5 (4) | C25—C24—O2—C23 | 115.2 (3) |
| C11—C12—C13—Cl1 | −178.78 (18) | C7—C8—Se1—N2 | −0.22 (17) |
| C12—C13—C14—C15 | −1.5 (4) | C9—C8—Se1—N2 | −175.37 (18) |
| Cl1—C13—C14—C15 | 178.69 (18) | N1—N2—Se1—C8 | 0.5 (2) |
| C13—C14—C15—C10 | 0.3 (3) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: NG5333).
References
- Bruker (2008). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Khanna, P. K. (2005). Phosphorus Sulfur Silicon Relat. Elem. 180, 951–955. [DOI] [PMC free article] [PubMed]
- Marx, A., Saravanan, S., Muthusubramanian, S., Manivannan, V. & Rath, N. P. (2008). Acta Cryst. E64, o349. [DOI] [PMC free article] [PubMed]
- Muthukumaran, J., Nachiappan, M., Chitra, S., Manisankar, P., Bhattacharya, S., Muthusubramanian, S., Krishna, R. & Jeyakanthan, J. (2011). Acta Cryst. E67, o2010–o2011. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813017790/ng5333sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813017790/ng5333Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813017790/ng5333Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



