Abstract
In the title molecule, C20H17Cl2NO2, the pyrrole moiety makes dihedral angles of 63.42 (11) and 70.43 (12)° with the chlorobenzene rings. The ethoxycarbonyl unit is present in a synperiplanar conformation with respect to the pyrrole ring, as indicated by the dihedral angle of 14.5 (3)°. In the crystal, molecules are linked into chains parallel to the a-axis direction by weak C—H⋯O hydrogen bonds.
Related literature
For the biological importance of pyrroles, see: Banwell et al. (2006 ▶); Mohamed et al. (2009 ▶); Sosa et al. (2002 ▶).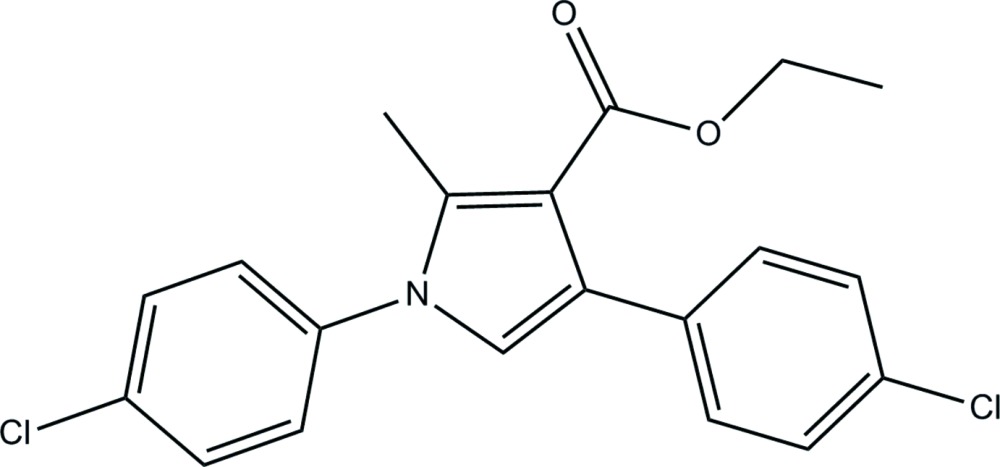
Experimental
Crystal data
C20H17Cl2NO2
M r = 374.25
Triclinic,
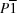
a = 8.037 (2) Å
b = 9.797 (3) Å
c = 12.510 (4) Å
α = 72.774 (16)°
β = 86.838 (16)°
γ = 76.804 (16)°
V = 915.9 (5) Å3
Z = 2
Mo Kα radiation
μ = 0.37 mm−1
T = 296 K
0.15 × 0.15 × 0.15 mm
Data collection
Bruker SMART APEXII CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2001 ▶) T min = 0.947, T max = 0.947
15843 measured reflections
4196 independent reflections
2759 reflections with I > 2σ(I)
R int = 0.032
Refinement
R[F 2 > 2σ(F 2)] = 0.044
wR(F 2) = 0.125
S = 1.03
4196 reflections
228 parameters
H-atom parameters constrained
Δρmax = 0.24 e Å−3
Δρmin = −0.29 e Å−3
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2009 ▶); software used to prepare material for publication: SHELXL97
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813019247/fb2292sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813019247/fb2292Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813019247/fb2292Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C2—H2⋯O8i | 0.93 | 2.58 | 3.453 (3) | 157 |
| C6—H6C⋯O8 | 0.96 | 2.42 | 3.041 (3) | 122 |
Symmetry code: (i)  .
.
Acknowledgments
C and KNN would like to thank the University of Mysore for awarding RFSMS fellowships [DV5/Physics/389/RFSMS/2009–2010/10.07.2012 and No. F4–1/2006(BSR)/7–131/2007, respectively].
supplementary crystallographic information
Comment
Pyrrole is a five-membered heterocyclic ring with one nitrogen atom. Its derivatives exhibit a variety of biological activities such as antitumor (Banwell et al., 2006) and antimicrobial (Mohamed et al., 2009) activities. They also inhibit protein kinase (Sosa et al., 2002). With this background of pyrrole derivatives, we have synthesized the title compound in order to study its crystal structure.
In the molecular structure of the title compound (Fig. 1), the dihedral angle between the pyrrole ring (N1/C2/C3/C4/C5) with phenyl rings (C19/C20/C21/C22/C23/C24) and (C12/C13/C14/C15/C16/C17) are 63.42 (11)° and 70.43 (12)°, respectively. The ethoxycarbonyl unit is in syn-periplanar conformation with respect to the pyrrole moiety, as indicated by the dihedral angle value of 14.5 (3)° (C3/C4/C7/O9). There are no classical hydrogen bonds and the crystal structure is stabilized by C—H···O hydrogen bonds only (see Table 1). C6—H6C···O8 forms an intramolecular hydrogen bond, while C2—H2···O8 links molecules which are parallel to the axis a. The packing of the molecules is shown in Fig. 2.
Experimental
To a stirred solution of para-chloroaniline (1.5 mmol), para-chlorobenzaldehyde (1.0 mmol) and ethyl acetoacetate (1.0 mmol) in nitromethane (1.5 ml), ferric chloride (FeCl3) (0.1 mmol) was added. The mixture was refluxed at 90–100°C for 6 hrs and then cooled to room temperature. The excess of solvent was removed under vacuum and the residue was directly purified by column chromatography using 60–120 silica gel with ethyl acetate in hexane (1:9) as eluent which afforded the desired product as yellow solid with 88% yield. The crude product has been recrystallized from hot ethanol. Typical size of the block-shaped crystals was 0.20 × 0.15 × 0.10 mm.
Refinement
All the H atoms were located in the difference electron density map. Nevertheless all the H atoms were situated into the idealized positions and allowed to ride on their parent atoms with C–H distances equal to 0.93, 0.96 and 0.97Å for aryl, methylene and methyl hydrogens. UisoHaryl/methylene = 1.2UeqCaryl/methylene and Umethyl = 1.5UeqCmethyl
Figures
Fig. 1.

The title molecule with the labelling scheme. The displacement ellipsoids are shown at the 50% probability level.
Fig. 2.
Packing diagram of the molecule viewed parallel to the a axis.
Crystal data
| C20H17Cl2NO2 | Z = 2 |
| Mr = 374.25 | F(000) = 388 |
| Triclinic, P1 | Dx = 1.357 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.037 (2) Å | Cell parameters from 4196 reflections |
| b = 9.797 (3) Å | θ = 1.7–27.5° |
| c = 12.510 (4) Å | µ = 0.37 mm−1 |
| α = 72.774 (16)° | T = 296 K |
| β = 86.838 (16)° | Block, yellow |
| γ = 76.804 (16)° | 0.15 × 0.15 × 0.15 mm |
| V = 915.9 (5) Å3 |
Data collection
| Bruker SMART APEXII CCD area-detector diffractometer | 4196 independent reflections |
| Radiation source: fine-focus sealed tube | 2759 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.032 |
| ω and φ scans | θmax = 27.5°, θmin = 1.7° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2001) | h = −10→10 |
| Tmin = 0.947, Tmax = 0.947 | k = −12→12 |
| 15843 measured reflections | l = −16→16 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.044 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.125 | H-atom parameters constrained |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0487P)2 + 0.2827P] where P = (Fo2 + 2Fc2)/3 |
| 4196 reflections | (Δ/σ)max < 0.001 |
| 228 parameters | Δρmax = 0.24 e Å−3 |
| 0 restraints | Δρmin = −0.29 e Å−3 |
| 66 constraints |
Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell e.s.d.'s are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement on F2 for ALL reflections except those flagged by the user for potential systematic errors. Weighted R-factors wR and all goodnesses of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The observed criterion of F2 > σ(F2) is used only for calculating -R-factor-obs etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl18 | 0.20768 (10) | 0.57041 (7) | 0.24828 (5) | 0.0884 (2) | |
| Cl25 | 0.78934 (10) | −0.46743 (9) | 1.16162 (6) | 0.1057 (3) | |
| O9 | −0.18070 (16) | 0.14269 (16) | 0.59033 (12) | 0.0585 (4) | |
| O8 | −0.27098 (18) | 0.0253 (2) | 0.75691 (13) | 0.0735 (5) | |
| N1 | 0.2589 (2) | −0.07110 (19) | 0.83555 (13) | 0.0521 (4) | |
| C13 | 0.1310 (3) | 0.2004 (2) | 0.46496 (17) | 0.0554 (5) | |
| H13 | 0.1005 | 0.1157 | 0.4616 | 0.067* | |
| C12 | 0.1613 (2) | 0.2151 (2) | 0.56891 (15) | 0.0458 (4) | |
| C3 | 0.1594 (2) | 0.0973 (2) | 0.67518 (15) | 0.0470 (4) | |
| C7 | −0.1558 (2) | 0.0648 (2) | 0.69825 (16) | 0.0498 (5) | |
| C4 | 0.0231 (2) | 0.0321 (2) | 0.73283 (15) | 0.0458 (4) | |
| C15 | 0.1876 (3) | 0.4336 (2) | 0.37081 (17) | 0.0562 (5) | |
| C17 | 0.2025 (3) | 0.3436 (2) | 0.56978 (17) | 0.0554 (5) | |
| H17 | 0.2219 | 0.3576 | 0.6380 | 0.066* | |
| C14 | 0.1448 (3) | 0.3078 (2) | 0.36672 (17) | 0.0589 (5) | |
| H14 | 0.1251 | 0.2949 | 0.2982 | 0.071* | |
| C19 | 0.3838 (2) | −0.1689 (2) | 0.91686 (16) | 0.0505 (5) | |
| C2 | 0.2999 (2) | 0.0304 (2) | 0.74178 (16) | 0.0533 (5) | |
| H2 | 0.4079 | 0.0502 | 0.7263 | 0.064* | |
| C22 | 0.6316 (3) | −0.3508 (3) | 1.06663 (17) | 0.0615 (6) | |
| C16 | 0.2157 (3) | 0.4528 (2) | 0.47182 (19) | 0.0640 (6) | |
| H16 | 0.2436 | 0.5388 | 0.4746 | 0.077* | |
| C6 | 0.0083 (3) | −0.1721 (3) | 0.92294 (19) | 0.0655 (6) | |
| H6A | −0.0078 | −0.1366 | 0.9874 | 0.098* | |
| H6B | 0.0816 | −0.2684 | 0.9426 | 0.098* | |
| H6C | −0.1003 | −0.1764 | 0.8973 | 0.098* | |
| C5 | 0.0887 (2) | −0.0707 (2) | 0.83166 (16) | 0.0501 (5) | |
| C23 | 0.6287 (3) | −0.3663 (3) | 0.9623 (2) | 0.0794 (8) | |
| H23 | 0.7100 | −0.4383 | 0.9421 | 0.095* | |
| C20 | 0.3894 (3) | −0.1548 (3) | 1.02199 (18) | 0.0664 (6) | |
| H20 | 0.3088 | −0.0826 | 1.0424 | 0.080* | |
| C21 | 0.5134 (3) | −0.2469 (3) | 1.09782 (18) | 0.0690 (6) | |
| H21 | 0.5162 | −0.2381 | 1.1697 | 0.083* | |
| C24 | 0.5039 (3) | −0.2739 (3) | 0.88682 (18) | 0.0721 (7) | |
| H24 | 0.5014 | −0.2832 | 0.8151 | 0.087* | |
| C10 | −0.3529 (3) | 0.1775 (3) | 0.54626 (19) | 0.0698 (6) | |
| H10A | −0.4272 | 0.2462 | 0.5798 | 0.084* | |
| H10B | −0.3972 | 0.0894 | 0.5630 | 0.084* | |
| C11 | −0.3464 (3) | 0.2438 (3) | 0.4224 (2) | 0.0822 (8) | |
| H11A | −0.3013 | 0.3303 | 0.4068 | 0.123* | |
| H11B | −0.4596 | 0.2694 | 0.3910 | 0.123* | |
| H11C | −0.2742 | 0.1744 | 0.3900 | 0.123* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl18 | 0.1196 (6) | 0.0733 (4) | 0.0583 (4) | −0.0226 (4) | 0.0054 (3) | 0.0019 (3) |
| Cl25 | 0.1053 (6) | 0.1152 (6) | 0.0738 (4) | 0.0282 (4) | −0.0484 (4) | −0.0242 (4) |
| O8 | 0.0469 (8) | 0.1037 (13) | 0.0608 (9) | −0.0179 (8) | 0.0010 (7) | −0.0098 (9) |
| O9 | 0.0441 (7) | 0.0719 (10) | 0.0534 (8) | −0.0092 (7) | −0.0091 (6) | −0.0103 (7) |
| N1 | 0.0445 (9) | 0.0604 (10) | 0.0448 (9) | −0.0098 (7) | −0.0067 (7) | −0.0056 (8) |
| C2 | 0.0458 (11) | 0.0623 (13) | 0.0474 (11) | −0.0157 (9) | −0.0044 (8) | −0.0057 (10) |
| C3 | 0.0447 (10) | 0.0518 (11) | 0.0444 (10) | −0.0085 (8) | −0.0044 (8) | −0.0146 (9) |
| C4 | 0.0431 (10) | 0.0495 (11) | 0.0440 (10) | −0.0073 (8) | −0.0011 (8) | −0.0143 (9) |
| C5 | 0.0445 (10) | 0.0551 (12) | 0.0495 (11) | −0.0099 (9) | −0.0014 (8) | −0.0141 (9) |
| C6 | 0.0567 (13) | 0.0673 (15) | 0.0614 (14) | −0.0137 (11) | 0.0008 (10) | −0.0022 (11) |
| C7 | 0.0471 (11) | 0.0542 (12) | 0.0470 (11) | −0.0065 (9) | −0.0031 (9) | −0.0163 (9) |
| C10 | 0.0461 (11) | 0.0895 (17) | 0.0670 (14) | −0.0050 (11) | −0.0159 (10) | −0.0172 (13) |
| C11 | 0.0730 (16) | 0.104 (2) | 0.0638 (15) | −0.0006 (14) | −0.0226 (12) | −0.0256 (14) |
| C12 | 0.0391 (9) | 0.0498 (11) | 0.0454 (10) | −0.0043 (8) | −0.0050 (7) | −0.0126 (9) |
| C13 | 0.0609 (12) | 0.0560 (12) | 0.0521 (12) | −0.0145 (10) | −0.0025 (9) | −0.0181 (10) |
| C14 | 0.0657 (13) | 0.0681 (14) | 0.0432 (11) | −0.0131 (11) | −0.0025 (9) | −0.0177 (10) |
| C15 | 0.0562 (12) | 0.0538 (12) | 0.0487 (11) | −0.0031 (9) | 0.0008 (9) | −0.0072 (10) |
| C16 | 0.0803 (15) | 0.0499 (12) | 0.0617 (14) | −0.0140 (11) | −0.0046 (11) | −0.0157 (11) |
| C17 | 0.0637 (13) | 0.0549 (13) | 0.0474 (11) | −0.0092 (10) | −0.0085 (9) | −0.0161 (10) |
| C19 | 0.0464 (10) | 0.0563 (12) | 0.0429 (10) | −0.0089 (9) | −0.0073 (8) | −0.0059 (9) |
| C20 | 0.0676 (14) | 0.0734 (15) | 0.0534 (13) | 0.0026 (11) | −0.0073 (10) | −0.0238 (11) |
| C21 | 0.0756 (15) | 0.0849 (17) | 0.0443 (11) | −0.0051 (13) | −0.0134 (10) | −0.0225 (12) |
| C22 | 0.0628 (13) | 0.0675 (14) | 0.0463 (11) | −0.0038 (11) | −0.0170 (9) | −0.0098 (10) |
| C23 | 0.0809 (16) | 0.0853 (18) | 0.0589 (14) | 0.0205 (13) | −0.0208 (12) | −0.0267 (13) |
| C24 | 0.0764 (15) | 0.0865 (17) | 0.0474 (12) | 0.0090 (13) | −0.0181 (10) | −0.0275 (12) |
Geometric parameters (Å, º)
| Cl18—C15 | 1.740 (2) | C19—C24 | 1.366 (3) |
| Cl25—C22 | 1.739 (3) | C20—C21 | 1.376 (3) |
| O8—C7 | 1.211 (2) | C21—C22 | 1.358 (4) |
| O9—C7 | 1.340 (2) | C22—C23 | 1.360 (3) |
| O9—C10 | 1.447 (3) | C23—C24 | 1.379 (4) |
| N1—C2 | 1.375 (3) | C2—H2 | 0.9300 |
| N1—C5 | 1.371 (2) | C6—H6A | 0.9600 |
| N1—C19 | 1.434 (3) | C6—H6B | 0.9600 |
| C2—C3 | 1.357 (3) | C6—H6C | 0.9600 |
| C3—C4 | 1.445 (2) | C10—H10A | 0.9700 |
| C3—C12 | 1.482 (3) | C10—H10B | 0.9700 |
| C4—C5 | 1.383 (3) | C11—H11A | 0.9600 |
| C4—C7 | 1.461 (2) | C11—H11B | 0.9600 |
| C5—C6 | 1.499 (3) | C11—H11C | 0.9600 |
| C10—C11 | 1.495 (3) | C13—H13 | 0.9300 |
| C12—C13 | 1.390 (3) | C14—H14 | 0.9300 |
| C12—C17 | 1.376 (3) | C16—H16 | 0.9300 |
| C13—C14 | 1.379 (3) | C17—H17 | 0.9300 |
| C14—C15 | 1.369 (3) | C20—H20 | 0.9300 |
| C15—C16 | 1.369 (3) | C21—H21 | 0.9300 |
| C16—C17 | 1.385 (3) | C23—H23 | 0.9300 |
| C19—C20 | 1.367 (3) | C24—H24 | 0.9300 |
| Cl18···C21i | 3.505 (3) | C10···H2v | 3.0500 |
| Cl18···H23ii | 3.0100 | C10···H16vii | 3.0400 |
| Cl25···H17iii | 3.0000 | C11···H16vii | 3.0700 |
| O8···C6 | 3.041 (3) | C15···H11Bx | 2.9100 |
| O8···C20iv | 3.377 (3) | C17···H10Ax | 2.9100 |
| O9···C12 | 2.971 (2) | C19···H6B | 2.7900 |
| O9···C13 | 2.957 (3) | H2···O8x | 2.5800 |
| O8···H10A | 2.7200 | H2···C10x | 3.0500 |
| O8···H10B | 2.5300 | H2···H10Bx | 2.5000 |
| O8···H20iv | 2.7200 | H6B···C19 | 2.7900 |
| O8···H21iv | 2.8500 | H6C···O8 | 2.4200 |
| O8···H2v | 2.5800 | H6C···C7 | 2.8500 |
| O8···H6C | 2.4200 | H10A···O8 | 2.7200 |
| O9···H13 | 2.7100 | H10A···C17v | 2.9100 |
| O9···H13vi | 2.7300 | H10B···O8 | 2.5300 |
| O9···H16vii | 2.9100 | H10B···H2v | 2.5000 |
| C6···O8 | 3.041 (3) | H11A···H16vii | 2.3500 |
| C6···C20 | 3.424 (4) | H11B···C15v | 2.9100 |
| C12···O9 | 2.971 (2) | H11B···H24vi | 2.5800 |
| C13···O9 | 2.957 (3) | H11C···C2vi | 3.0000 |
| C15···C17vii | 3.567 (3) | H11C···C3vi | 2.9500 |
| C16···C17vii | 3.473 (3) | H13···O9 | 2.7100 |
| C16···C16vii | 3.468 (4) | H13···O9vi | 2.7300 |
| C17···C15vii | 3.567 (3) | H13···C7vi | 2.9900 |
| C17···C16vii | 3.473 (3) | H16···O9vii | 2.9100 |
| C20···O8iv | 3.377 (3) | H16···C10vii | 3.0400 |
| C20···C6 | 3.424 (4) | H16···C11vii | 3.0700 |
| C21···Cl18viii | 3.505 (3) | H16···H11Avii | 2.3500 |
| C23···C23ix | 3.582 (4) | H17···C2 | 3.0100 |
| C2···H11Cvi | 3.0000 | H17···Cl25iii | 3.0000 |
| C2···H24 | 3.0200 | H20···O8iv | 2.7200 |
| C2···H17 | 3.0100 | H21···O8iv | 2.8500 |
| C3···H11Cvi | 2.9500 | H23···Cl18ii | 3.0100 |
| C7···H13vi | 2.9900 | H24···C2 | 3.0200 |
| C7···H6C | 2.8500 | H24···H11Bvi | 2.5800 |
| C7—O9—C10 | 116.77 (15) | C19—C24—C23 | 120.4 (2) |
| C2—N1—C5 | 109.47 (16) | N1—C2—H2 | 125.00 |
| C2—N1—C19 | 122.99 (16) | C3—C2—H2 | 125.00 |
| C5—N1—C19 | 127.29 (16) | C5—C6—H6A | 109.00 |
| N1—C2—C3 | 109.90 (16) | C5—C6—H6B | 109.00 |
| C2—C3—C4 | 105.42 (16) | C5—C6—H6C | 109.00 |
| C2—C3—C12 | 122.85 (16) | H6A—C6—H6B | 109.00 |
| C4—C3—C12 | 131.70 (16) | H6A—C6—H6C | 110.00 |
| C3—C4—C5 | 108.37 (15) | H6B—C6—H6C | 109.00 |
| C3—C4—C7 | 128.27 (17) | O9—C10—H10A | 110.00 |
| C5—C4—C7 | 123.36 (16) | O9—C10—H10B | 110.00 |
| N1—C5—C4 | 106.84 (16) | C11—C10—H10A | 110.00 |
| N1—C5—C6 | 121.23 (18) | C11—C10—H10B | 110.00 |
| C4—C5—C6 | 131.92 (17) | H10A—C10—H10B | 108.00 |
| O8—C7—O9 | 122.15 (17) | C10—C11—H11A | 109.00 |
| O8—C7—C4 | 125.71 (18) | C10—C11—H11B | 109.00 |
| O9—C7—C4 | 112.13 (15) | C10—C11—H11C | 109.00 |
| O9—C10—C11 | 107.67 (19) | H11A—C11—H11B | 109.00 |
| C3—C12—C13 | 123.03 (18) | H11A—C11—H11C | 110.00 |
| C3—C12—C17 | 119.97 (17) | H11B—C11—H11C | 110.00 |
| C13—C12—C17 | 116.92 (18) | C12—C13—H13 | 119.00 |
| C12—C13—C14 | 121.90 (19) | C14—C13—H13 | 119.00 |
| C13—C14—C15 | 119.53 (19) | C13—C14—H14 | 120.00 |
| Cl18—C15—C14 | 120.60 (16) | C15—C14—H14 | 120.00 |
| Cl18—C15—C16 | 119.25 (17) | C15—C16—H16 | 120.00 |
| C14—C15—C16 | 120.15 (19) | C17—C16—H16 | 120.00 |
| C15—C16—C17 | 119.7 (2) | C12—C17—H17 | 119.00 |
| C12—C17—C16 | 121.80 (19) | C16—C17—H17 | 119.00 |
| N1—C19—C20 | 121.23 (19) | C19—C20—H20 | 120.00 |
| N1—C19—C24 | 119.19 (18) | C21—C20—H20 | 120.00 |
| C20—C19—C24 | 119.5 (2) | C20—C21—H21 | 120.00 |
| C19—C20—C21 | 120.4 (2) | C22—C21—H21 | 120.00 |
| C20—C21—C22 | 119.3 (2) | C22—C23—H23 | 120.00 |
| Cl25—C22—C21 | 119.70 (17) | C24—C23—H23 | 120.00 |
| Cl25—C22—C23 | 119.1 (2) | C19—C24—H24 | 120.00 |
| C21—C22—C23 | 121.2 (2) | C23—C24—H24 | 120.00 |
| C22—C23—C24 | 119.2 (3) | ||
| C10—O9—C7—O8 | −0.2 (3) | C7—C4—C5—C6 | 1.5 (4) |
| C10—O9—C7—C4 | 178.21 (19) | C3—C4—C7—O8 | −167.2 (2) |
| C7—O9—C10—C11 | −171.6 (2) | C3—C4—C7—O9 | 14.5 (3) |
| C5—N1—C2—C3 | −0.5 (2) | C5—C4—C7—O8 | 12.2 (3) |
| C19—N1—C2—C3 | 174.18 (18) | C5—C4—C7—O9 | −166.22 (19) |
| C2—N1—C5—C4 | 0.6 (2) | C3—C12—C13—C14 | −175.6 (2) |
| C2—N1—C5—C6 | 179.4 (2) | C17—C12—C13—C14 | 1.3 (3) |
| C19—N1—C5—C4 | −173.79 (19) | C3—C12—C17—C16 | 176.1 (2) |
| C19—N1—C5—C6 | 5.0 (3) | C13—C12—C17—C16 | −0.9 (3) |
| C2—N1—C19—C20 | 110.8 (2) | C12—C13—C14—C15 | −0.8 (4) |
| C2—N1—C19—C24 | −66.3 (3) | C13—C14—C15—Cl18 | 179.48 (19) |
| C5—N1—C19—C20 | −75.4 (3) | C13—C14—C15—C16 | −0.3 (4) |
| C5—N1—C19—C24 | 107.5 (2) | Cl18—C15—C16—C17 | −179.05 (19) |
| N1—C2—C3—C4 | 0.2 (2) | C14—C15—C16—C17 | 0.7 (4) |
| N1—C2—C3—C12 | 178.42 (18) | C15—C16—C17—C12 | −0.1 (4) |
| C2—C3—C4—C5 | 0.2 (2) | N1—C19—C20—C21 | −178.0 (2) |
| C2—C3—C4—C7 | 179.6 (2) | C24—C19—C20—C21 | −1.0 (4) |
| C12—C3—C4—C5 | −177.8 (2) | N1—C19—C24—C23 | 177.9 (2) |
| C12—C3—C4—C7 | 1.6 (4) | C20—C19—C24—C23 | 0.8 (4) |
| C2—C3—C12—C13 | 116.0 (2) | C19—C20—C21—C22 | 0.9 (4) |
| C2—C3—C12—C17 | −60.8 (3) | C20—C21—C22—Cl25 | 179.8 (2) |
| C4—C3—C12—C13 | −66.3 (3) | C20—C21—C22—C23 | −0.6 (4) |
| C4—C3—C12—C17 | 116.9 (2) | Cl25—C22—C23—C24 | −179.9 (2) |
| C3—C4—C5—N1 | −0.5 (2) | C21—C22—C23—C24 | 0.5 (4) |
| C3—C4—C5—C6 | −179.1 (2) | C22—C23—C24—C19 | −0.6 (4) |
| C7—C4—C5—N1 | −179.95 (18) |
Symmetry codes: (i) x, y+1, z−1; (ii) −x+1, −y, −z+1; (iii) −x+1, −y, −z+2; (iv) −x, −y, −z+2; (v) x−1, y, z; (vi) −x, −y, −z+1; (vii) −x, −y+1, −z+1; (viii) x, y−1, z+1; (ix) −x+1, −y−1, −z+2; (x) x+1, y, z.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C2—H2···O8x | 0.93 | 2.58 | 3.453 (3) | 157 |
| C6—H6C···O8 | 0.96 | 2.42 | 3.041 (3) | 122 |
Symmetry code: (x) x+1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: FB2292).
References
- Banwell, M. G., Hamel, E., Hockless, D. C. R., Verdier-Pinard, P., Willis, A. C. & Wong, D. J. (2006). Bioorg. Med. Chem. 14, 4627–4638. [DOI] [PubMed]
- Bruker (2009). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Mohamed, M. S., El-Domany, R. A. & El-Hameed, R. H. A. (2009). Acta Pharm. 59, 145–158. [DOI] [PubMed]
- Sheldrick, G. M. (2001). SADABS, University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sosa, A. C. B., Yakushijin, K. & Horne, D. A. (2002). J. Org. Chem. 67, 4498–4500. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813019247/fb2292sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813019247/fb2292Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813019247/fb2292Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



