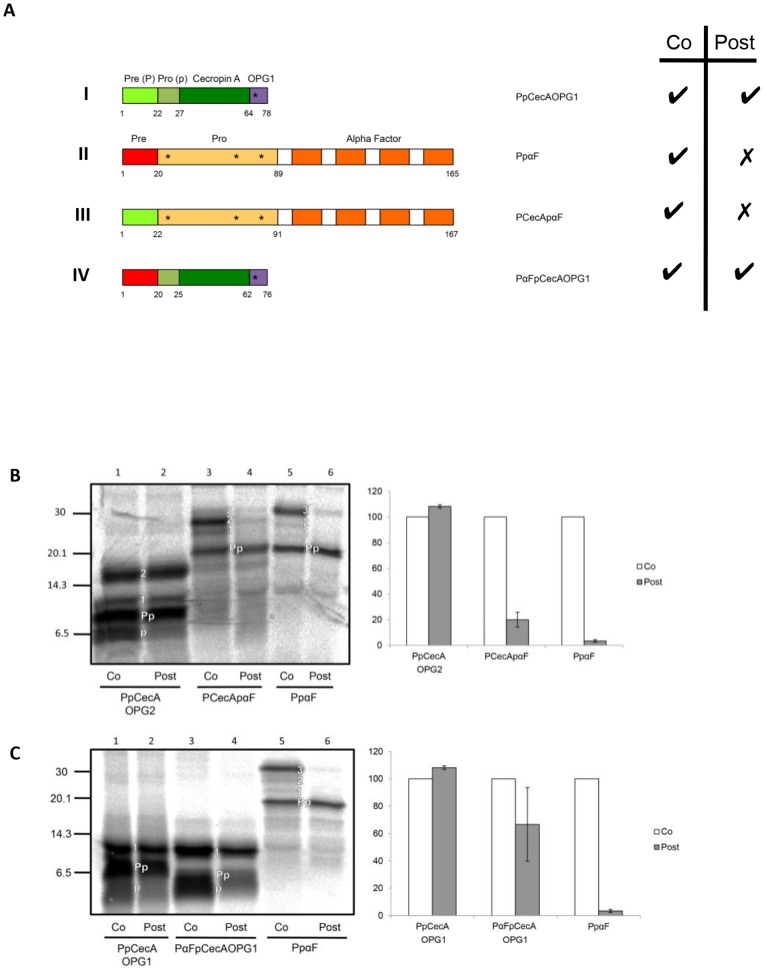
Figure 1. Signal sequence chimeras of PpCecA and PpαF.
A. Schematic representation of proteins derived from PpCecA and PpαF. Signal sequences (denoted Pre or P) are in light green and red respectively, whilst the purple region denotes an opsin epitope tag bearing one N-glycosylation site (OPG1). The OPG2 tag contains an additional four residues that includes a second N-glycosylation site, and both PpCecAOPG1 and PpCecAOPG2 are as previously described [16]. Numbers indicate first and last amino acids of each region, and asterisks indicate the approximate location of sites for N-linked glycosylation. The ability for each protein to exploit co- and post-translational translocation pathways, as deduced from the data shown in panels B and C, is indicated by a tick. B. The PpCecA signal sequence is not sufficient to enable efficient post-translational translocation of PpαF. The precursors indicated were synthesized in vitro using rabbit reticulocyte lysate and [35S] methionine in either the presence of canine pancreatic microsomes (co) or prior to their addition (post). The membrane fraction was isolated by centrifugation and associated proteins analyzed by SDS-PAGE and phosphorimaging. The untranslocated (Pp), singly, doubly and triply N-glycosylated (1, 2, 3) and signal cleaved (p) forms of the precursors are indicated (see also Figs. S1 and S2). The amount of post-translationally translocated material as a proportion of the total membrane associated material was quantified and is expressed relative to the proportion of material translocated under co-translational conditions (set to 100%). Results are given as means (±s.e) for n = 3. C. PpCecA can efficiently translocate post-translationally with the PpαF signal sequence. The precursors indicated were synthesized and the resulting data analyzed as described for B, with results given as means (±s.e) for n = 3.