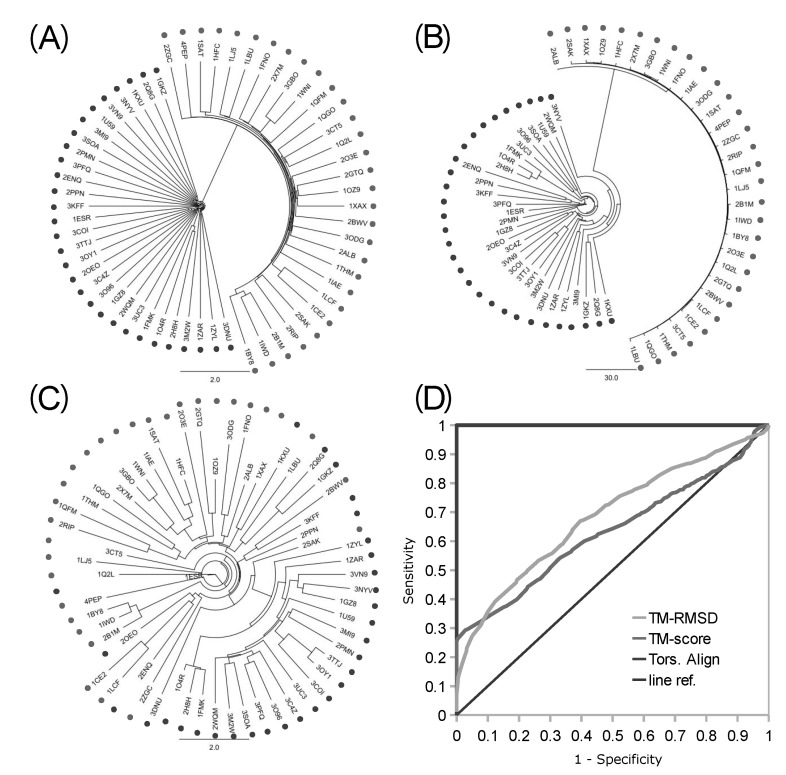
Fig. 4.
Clustering trees and accuracy analysis from intermingled kinases and proteases. The clustering trees of 30 kinases (dark dots) and 30 proteases (pale dots) built with logPr (A), RamRMSD (B), and TM-align RMSD (C) are displayed. Backbone torsion angle methods showed perfect delineation of kinases and proteases, while TM-align showed rather promiscuous clustering, with partially correct clusters of kinases. Only the result of the RMSD of TM-align was displayed, which showed better accuracy than the TM-score result. Receiver operating characteristic (ROC) curve (D) shows perfect performance of the backbone torsion angle alignment with area under the ROC curve (AUROC) of 1.0, while the TM-align method showed similar performance with an AUROC of 0.6846 (RMSD) and 0.6319 (TM-score) to that of a previous set of little functional difference [1].