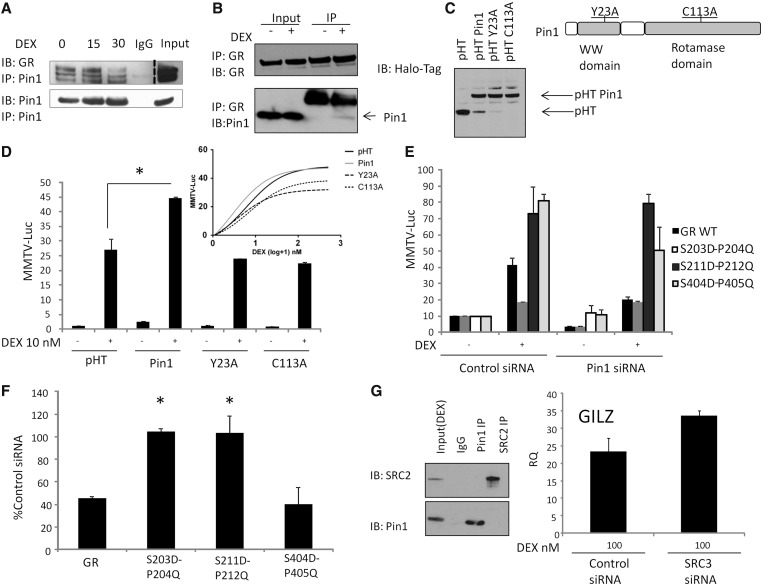
Figure 5.
Pin1 interactions with the GR. (A) A549 cells were stimulated with DEX (100 nM) for the indicated time, Pin1 was immunoprecipitated from cell lysates and immunoblots probed for GR (dashed line represents a different exposure for input). (B) Reciprocal immunoprecipitaions for GR. Seven percent of the input sample was used in the immunoblot. (C) Expression of Halo-Pin1 mutants and their location in Pin1. pHalo (pHT), pHT-Pin1 WT, pHT-Pin1 Y23A and pHT-Pin1 C113A expression in A549 cells. (D) A549 cells were transfected with WT Pin1, Pin1 Y23A, Pin1 C113A and MMTV-luc for 16 h, after which DEX 1-500 nM was added for 6 h. (E) A549 cells were transfected with Pin1 siRNA for 48 h, after 24 h cells were transfected with a MMTV-luciferase reporter. In all, 10 nM DEX was added for 6 h (n = 6). (F) HEK293T cells were transfected with control or Pin1 siRNA for 16 h followed by transfection with MMTV-luciferase reporter and Halo-tag GR WT, S203D-P204Q, S211D-P212Q or S404D-P405Q. Cells were then pre-treated with 10 nM DEX for 6 h. Graphs show mean (±SD) of the normalized fold change in MMTV promoter activity (n = 3). (G) A549 cells were treated with DEX for 1 h, cell lysates were prepared and SRC-2 or Pin1 immunoprecipitated and immunobloted with either SRC-2 or Pin1 antibodies. A549 cells were treated with SRC-3 siRNA for 48 h and treated with DEX for 2 h before the levels of the GILZ gene were determined by qPCR (n = 3). Statistical analysis was determined using a one-way ANOVA and a Bonferroni post-hoc test (P < 0.05*).