Figure 1.
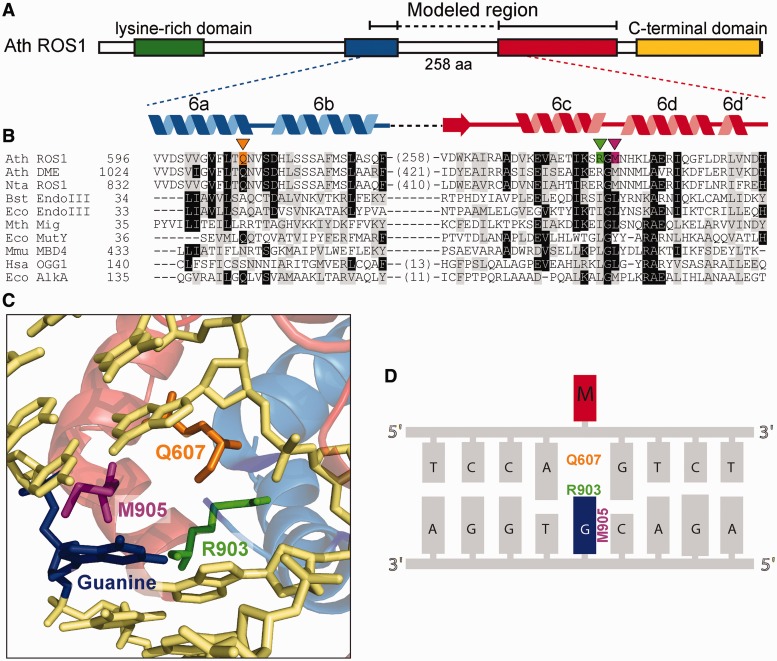
Identification of putative helix-invading residues in ROS1. (A) Schematic diagram showing conserved regions among members of the ROS1/DME family: a N-terminal lysine-rich region (green), a noncontiguous DNA glycosylase domain distributed over two segments (blue and red) separated by a nonstructured linker region and a highly conserved C-terminal domain (yellow) that is not found in any other protein family. (B) Multiple sequence alignment of part of the DNA glycosylase domain of ROS1/DME proteins and several HhH-GPD superfamily members. ROS1 amino acids analyzed in this work are indicated by inverted triangles and highlighted in orange (Q607), green (R903) or pink (M905). Names of organisms are abbreviated as follows: Ath, Arabidopsis thaliana; Nta, Nicotiana tabacum; Bst, Bacillus stearothermophilus; Eco, Escherichia coli; Mth, Methanobacterium thermoautotrophicum; Mmu, Mus musculus; Hsa, Homo sapiens. Genbank accession numbers are as follows: Ath ROS1: AAP37178; Ath DME: ABC61677; Nta ROS1: BAF52855; Bst EndoIII: 1P59; Eco EndoIII: P20625; Mth Mig: NP_039762; Eco MutY: NP_417436; Mmu MBD4: 1NGN; Hsa OGG1: O15527; Eco AlkA: P04395. (C) Structural model for the DNA glycosylase domain of ROS1 bound to a DNA containing an abasic site. The position of Q607, R903 and M905 residues (colored as in panel B) in relation to the estranged guanine (blue) are shown. The model was generated as described in (20) and the figure was prepared with PyMOL (http://www.pymol.org). (D) Schematic sequence diagram indicating the predicted interactions between mutated amino acids and the orphan guanine (blue) opposite 5-meC (M, in red).