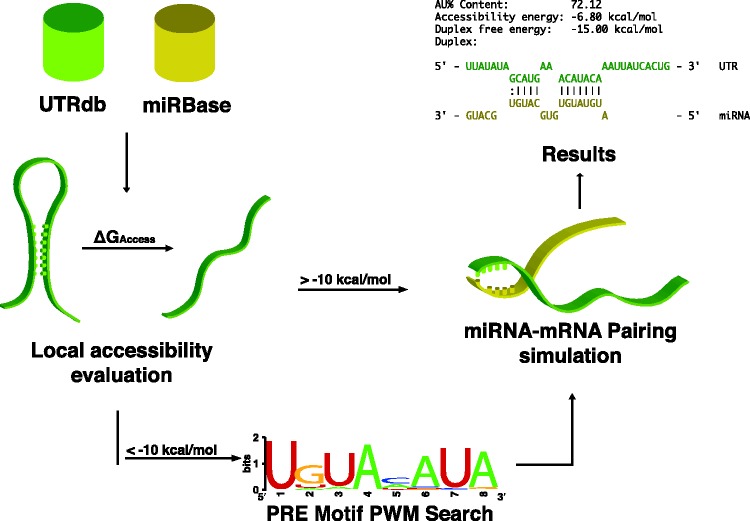
Figure 2.
Schematic representation of MREdictor’s pipeline. Given a target to test and a miRNA, MREdictor obtains sequences for the 3′-UTR from UTRdb and for the miRNA from miRBase. For every possible seed-match, the local accessibility is evaluated, and regions exceeding the ΔGaccess energy cost of −10 kcal/mol are subjected to a Positional Weight Matrix scan for possible PRE motifs. If no PRE is discovered, the site is discarded. Sites that pass this first filtering step are then subjected to a simulation of duplex formation and are filtered according to their free energy (ΔGduplex).