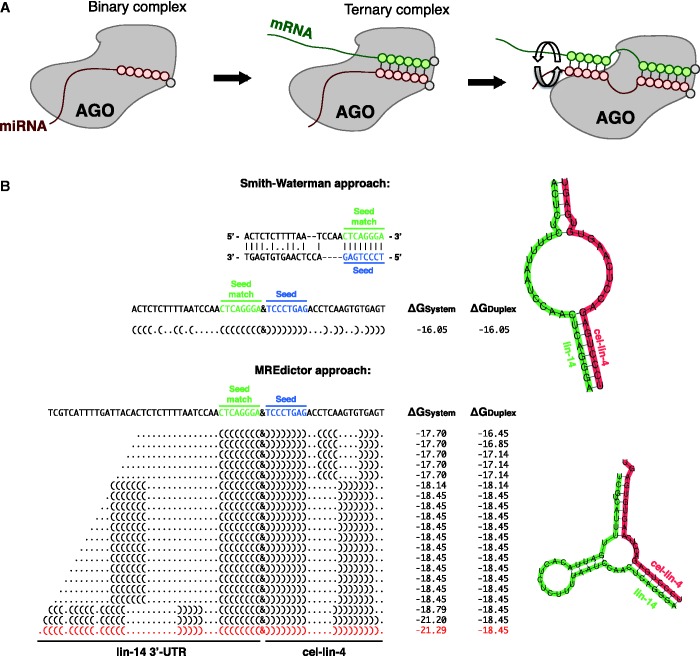
Figure 3.
Two step calculation of miRNA-MRE pairing. (A) Two-step model of the miRNA–mRNA interaction. (B) Comparison of a Smith–Waterman-like approach and the MREdictor method for miRNA-MRE duplex modeling, which consist of the nucleation of the miRNA-MRE duplex at the seed level followed by an extended 3′-end pairing (see ‘Materials and Methods’ section), applied to a well-known worm MRE. Previous studies have shown that, in Caenorhabditis elegans, the lin-4 miRNA can target lin-14, forming a bulged structure within lin-14 3′-UTR (25,33,34). A Smith–Waterman-like approach tends to maximize only the number of interactions between the miRNA and the target, minimizing the number of gaps and the extension of the gaps; thus, it fails to correctly predict the duplex secondary structure and underestimates the duplex free energy.