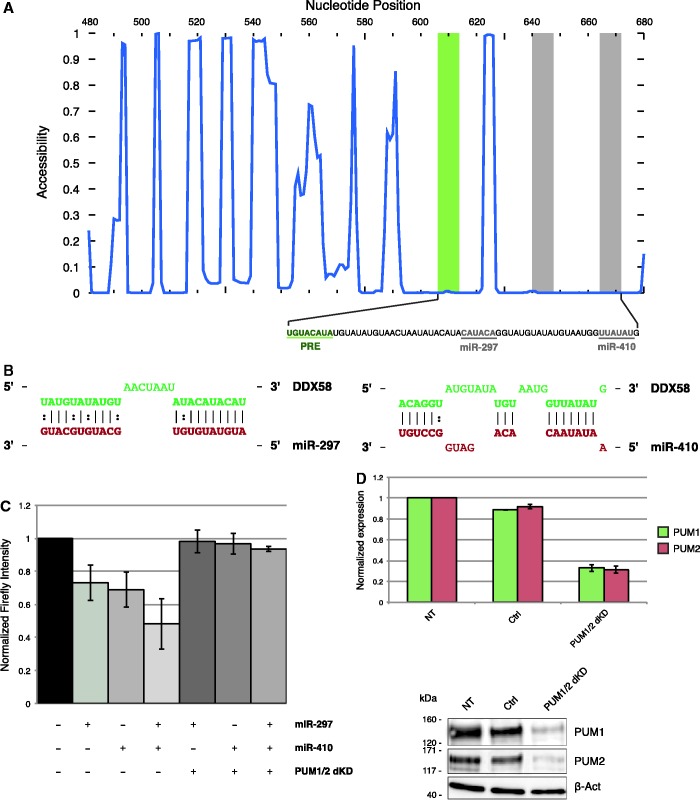
Figure 5.
Validation of PUM-dependent sites predicted by MREdictor. (A) Accessibility plot of a DDX58 3′-UTR window containing two MREs for miR-297 and miR-410, which are predicted to be dependent on Pumilio proteins binding to their cognate PRE motif. (B) Schematic representation of the two MREs, as predicted by MREdictor. (C) Dual luciferase assay validation of the two predicted MREs within DDX58 3′-UTR. Data are averaged over four replicates, and error bars are given for S.Ds. (D) RT-qPCR analysis and western blot of HEK293 FT cells transfected either with a negative control or two siRNAs targeting PUM1 and PUM2.