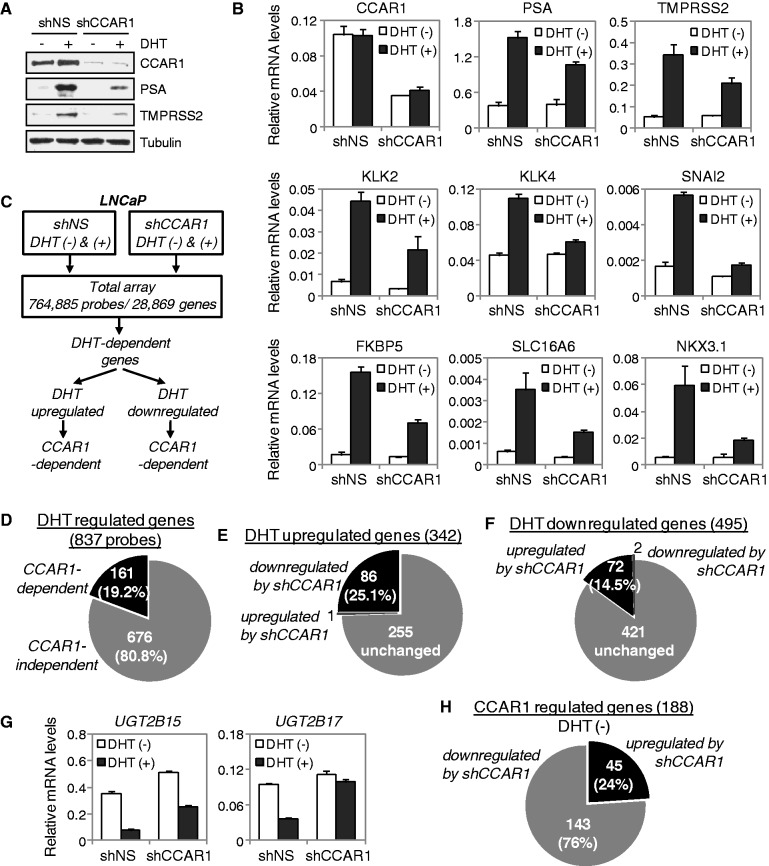
Figure 2.
Requirement of CCAR1 for AR function and identification of CCAR1-dependent target genes by microarray analysis. (A and B) LNCaP cells infected with lentiviruses encoding a non-specific (shNS) or CCAR1 shRNA (shCCAR1) were treated with 10 nM DHT or ethanol vehicle for 24 h. Protein levels were monitored by immunoblot using the indicated antibodies (A). Total RNA was examined by real-time qRT-PCR analysis with primers specific for the indicated mRNAs (B). Results shown were normalized to β-actin mRNA levels and are means ± SD (n = 3). (C) Flow chart depicting the strategy of cDNA microarray analysis and CCAR1-dependent gene selection process. (D) CCAR1-dependent genes represent 19.2% of all differentially expressed genes by DHT. (E) Pie graph shows that among all DHT-upregulated genes, 25.1% of them are downregulated by CCAR1 depletion. (F) Pie graph shows that among all DHT-downregulated genes, 14.5% of them are upregulated by CCAR1 depletion. (G) CCAR1 is required for androgen-repressed expression of UGT2B15 and UGT2B17 genes. LNCaP cells expressing shNS or shCCAR1 were treated with 10 nM DHT or ethanol vehicle for 24 h. The indicated mRNA levels were determined by qRT-PCR as described in (B). (H) Pie graph shows that 188 genes are differentially expressed by CCAR1 depletion in the absence of DHT.