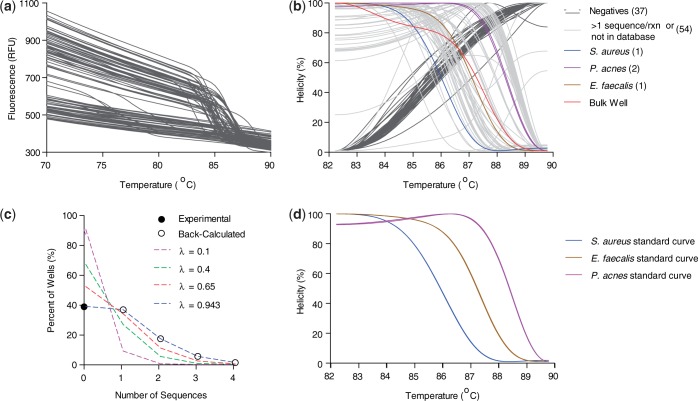
Figure 6.
A heterogeneous mixture of bacterial gDNA is resolvable by U-dHRM. (a) Raw dHRM data showing the loss of fluorescence in all but one well of a 96-well plate across which a dilute mixture of S. aureus, E. faecalis and P. acnes was dispersed. (b) Normalized, temperature calibrated and database matched digital melt curves after 70 cycles of PCR show broad-based amplified, single copy detection of each of the four input bacteria (colored melt curves), detection of wells containing multiple copies of input or contaminating gDNA templates from Taq polymerase (light gray, non-matching curves), and negative wells (dark gray melt curves). The number of copies detected is shown in parentheses next to each legend label. The corresponding ‘bulk’ well (red curve) where an amount of each target gDNA equivalent to that diluted across the rest of the plate, and also including contaminating Taq gDNA, was assayed by conventional bulk HRMA. (c) Poisson distribution matching to the results of broad-based digital detection of polymicrobial input by U-dHRM. Enumeration of negatives allows us to calculate that an experimental occupancy of ∼0.943 was achieved. (d) Standard database curves used to identify target melt curves.