Figure 7.
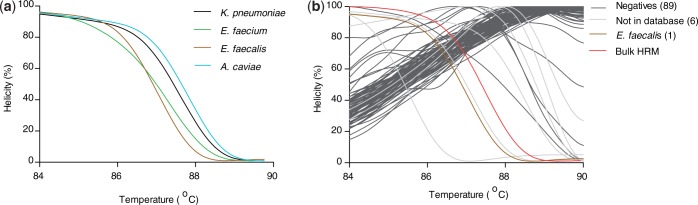
U-dHRM identifies bacteria in a clinical blood bottle. (a) Standard database melt curves generated from four types of bacteria isolated from the polymicrobial blood bottle. (b) Normalized, temperature calibrated and database matched digital melt curves after 70 cycles of PCR. Broad-based amplified, single copy detection of E. faecalis in the blood sample extract was achieved (brown melt curve). The number of wells containing single contaminating gDNA templates from Taq polymerase (light gray non-matching curves) is few. Negative wells (dark gray melt curves) give a calculated λ = 0.075. The number of copies detected is shown in parentheses next to each legend label. The corresponding ‘bulk’ melt curve (red curve) where an amount of DNA sample equivalent to that diluted across the rest of the plate, and also including contaminating Taq gDNA, was assayed by conventional bulk HRMA.
