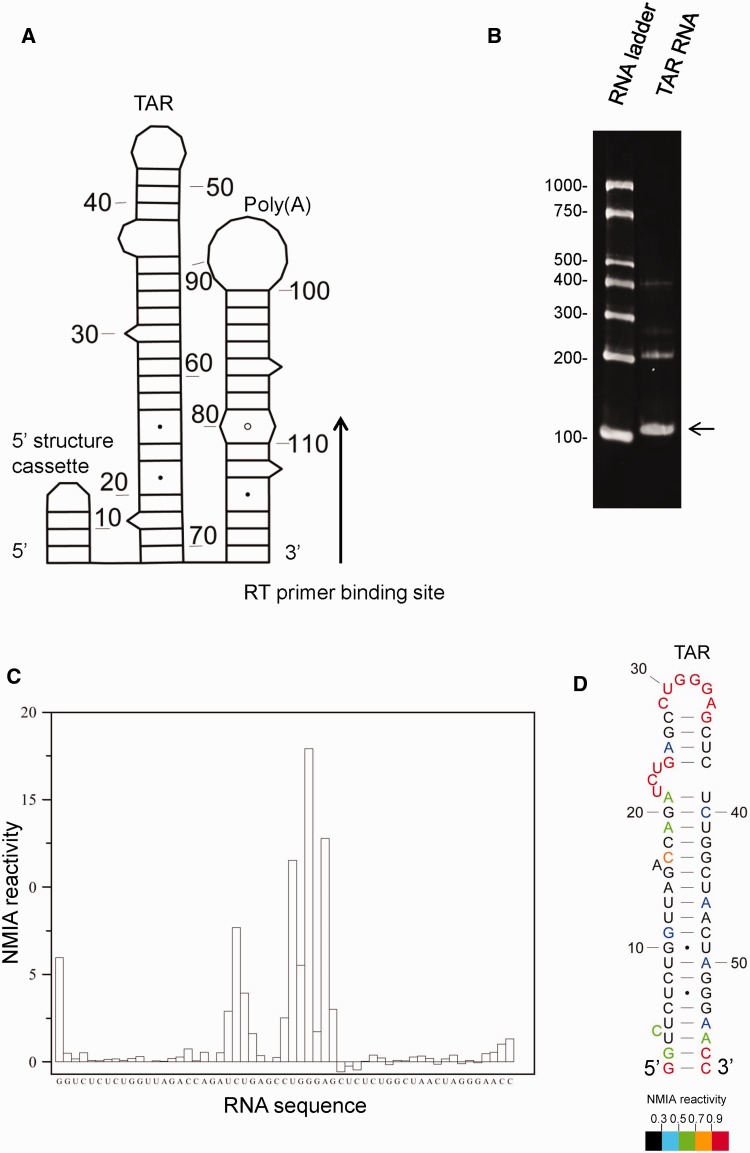
Figure 1.
In-gel SHAPE probing of a well-characterized stable RNA structure generates accurate reproducible results. (A) HIV-1 TAR RNA was appended with a 5′ structure cassette and poly(A) stem-loop. Nucleotides are numbered every 10 bases. (B) Ethidium bromide-stained native polyacrylamide gel. Arrow shows the monomeric TAR band that was excised for in-gel structural analysis. (C) NMIA reactivity trace of the in-gel probed monomeric TAR RNA. (D) RNAstructure prediction of TAR RNA, using in-gel SHAPE data as pseudo-free energy constraints. Individual NMIA reactivities are shown using different colours. Nucleotides are numbered every 10 bases.