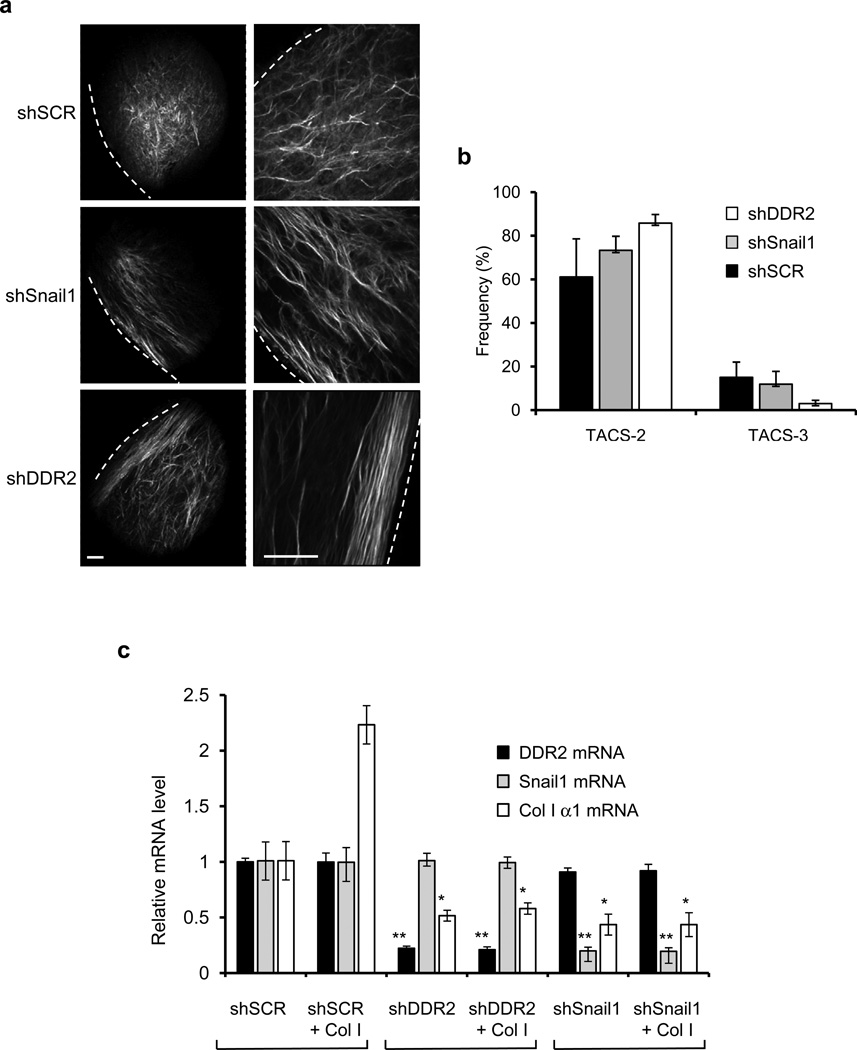
Figure 4. Collagen fiber alignment at tumor-ECM boundary.
(a) Second harmonic generation imaging of collagen fibers at the tumor-ECM boundary of primary tumors from xenografts of 4T1 cells transduced with control shSCR, shSnail1, or shDDR2 RNAi expressing lentiviruses. Dashed line represents the tumor boundary. Scale bars = 100µm. Both magnifications are of the same tumor but at different locations. (b) Using CurvAlign software, the angle of collagen fibers relative to the tumor boundary was determined. Each column consists of n=3 tumors with three fields of view for each (nine images for each tumor). All angles from 0–30 degrees were catagorized as “TACS-2”, parallel to the tumor boundary. Fibers in the range of 60–90 degrees were categorized as “TACS-3”, perpendicular to the tumor boundary. Means and S.E.M. are shown for one experiment. Primary data are provided in the Statistics Source Data file in the supplementary information. (c) MDA-MB-231 cells shRNAi-depleted of Snail1 (shSnail1), DDR2 (shDDR2), or control (SCR) were added to uncoated plates or plates coated with 2 mg/ml of collagen I for 8 hours and Q-PCR determination of mRNA levels for collagen I (black columns), Snail1 (light gray columns), and DDR2 (dark gray columns) relative to control cells not added to collagen-coated plates was determined. Means and s.d. for three independent experiments; three wells of cells were analyzed each experiment. P-values calculated using unpaired, two-sided Student’s t-tests. * P<0.05; ** P<0.01.