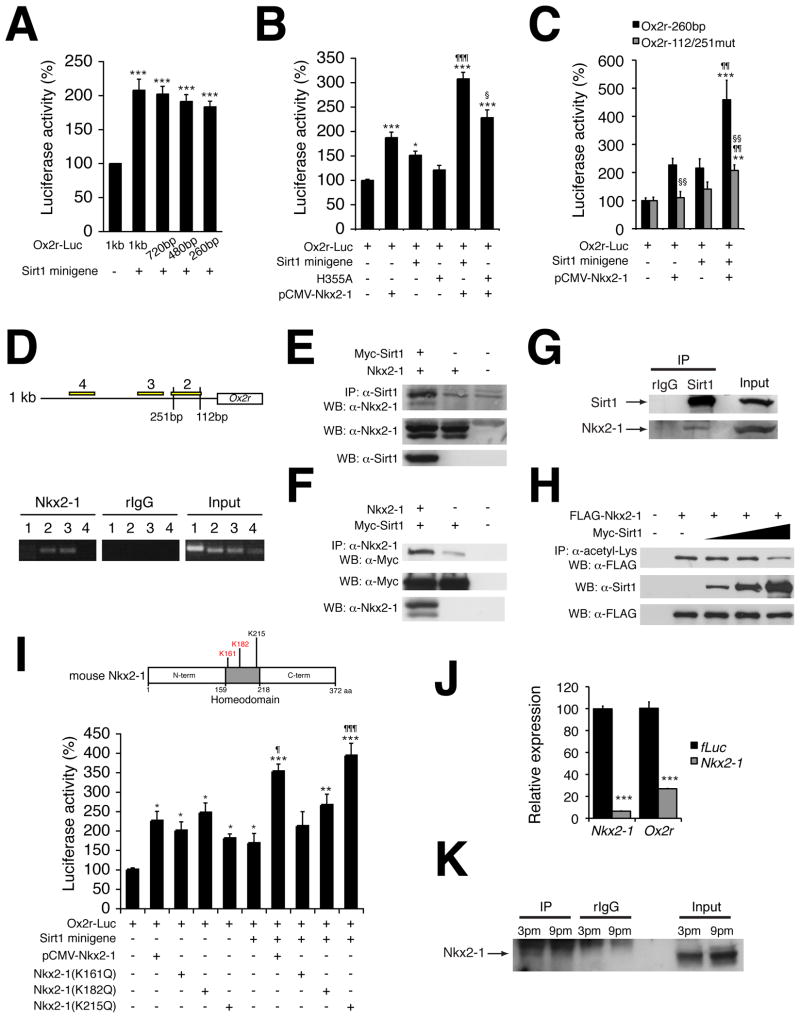
Figure 4. Sirt1 enhances Ox2r promoter activity by interacting with and deacetylating a homeodomain transcription factor Nkx2-1.
(A–C) Luciferase activities driven by the 1-kb and truncated Ox2r promoter fragments (A and B, Ox2r-Luc) or the 260-bp Ox2r promoter or mutant carrying mutations in two Nk2 family binding sites (C, Ox2r-112/251mut) in HEK293 cells co-transfected with a Sirt1 minigene or mutant Sirt1 (H355A) (A and B) and/or Nkx2-1 (pCMV-Nkx2-1) (B and C). Luciferase activities in cells co-transfected with a backbone vector were normalized to 100%. Results are shown as mean values ± S.E. [control vs treatment, *p<0.05, **p<0.01, ***p<0.001 by one-way ANOVA with Tukey-Kramer post hoc test (A–C); pCMV-Nkx2-1 alone vs pCMV-Nkx2-1 with Sirt1 minigene, ¶p<0.05, 1¶¶p<0.01, ¶¶¶p<0.001 by Student’s t-test (B and C); pCMV-Nkx2-1 with Sirt1 minigene vs pCMV-Nkx2-1 with H355A (B) or Ox2r-260bp vs Ox2r-112/251mut (C), §p<0.05, §§p<0.01 by Student’s t-test, n=3–9]. (D) Chromatin-immunoprecipitation for Nkx2-1 in the hypothalamus. Primer set 1 is designed for an unrelated genomic region. Locations of primer sets 2–4 are shown in the upper panel. Nk2 family binding sites are located at −112 and −251 positions with respect to the transcription start site in the Ox2r promoter. (E–G) Co-immunoprecipitation showing the interaction of Sirt1 and Nkx2-1 overexpressed in HEK293 cells (E and F) and in the hypothalamic extracts (G). (H) Deacetylation of Nkx2-1 by Sirt1 in HEK293 cells. (I) Luciferase activity of Ox2r-Luc in HEK293 cells co-transfected with a Sirt1 minigene and/or pCMV-Nkx2-1 or Nkx2-1 mutants carrying different point mutations (K161Q, K182Q, or K215Q). Results are shown as mean values ± S.E. (control vs treatment, *p<0.05, **p<0.01, ***p<0.001 by one-way ANOVA with Tukey-Kramer post hoc test; pCMV-Nkx2-1 alone vs pCMV-Nkx2-1 with Sirt1 minigene or pCMV-Nkx2-1 (K215Q) alone vs pCMV-Nkx2-1 (K215Q) with Sirt1 minigene, ¶p<0.05, ¶¶¶p<0.001 by Student’s t-test, n=6). These three lysines lie within the homeodomain of Nkx2-1 protein as shown in the schematic structure of Nkx2-1. Acetylated lysine residues are shown in red. (J) mRNA expression levels of Nkx2-1 and Ox2r after knockdown of Nkx2-1 in primary hypothalamic neurons. mRNA expression levels were normalized to those in control firefly luciferase (fLuc) knockdown cells for each gene. Results are shown as mean value ± S.E. (control vs treatment, ***p<0.001 by Student’s t-test, n=3). (K) Acetylation levels of Nkx2-1 in hypothalamic extracts at 3pm (light time) and 9pm (dark time). See also Figure S4.