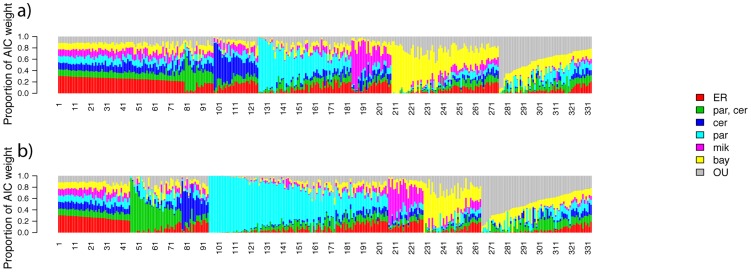
Figure 3. Inference of regulatory evolution in yeast pathways from experimental expression measurements.
Each panel reports results of phylogenetic inference of evolutionary histories of gene expression change from one set of experimental transcriptional profiling data. In a given panel, each vertical bar reports results of maximum-likelihood fits of Brownian-motion and Ornstein-Uhlenbeck models to expression of the genes of one Gene Ontology process term; the total proportion of a bar corresponding to a particular color indicates the Akaike weight of the corresponding model (legend at right, with labels as in Figure 1). Bars are sorted by the model with maximum Akaike weight. (a), Inference of cis-regulatory variation from interspecies hybrids; numerical indices correspond to rows in Table S3. (b), Inference from measurements of total expression in species homozygotes; numerical indices correspond to rows in Table S4.