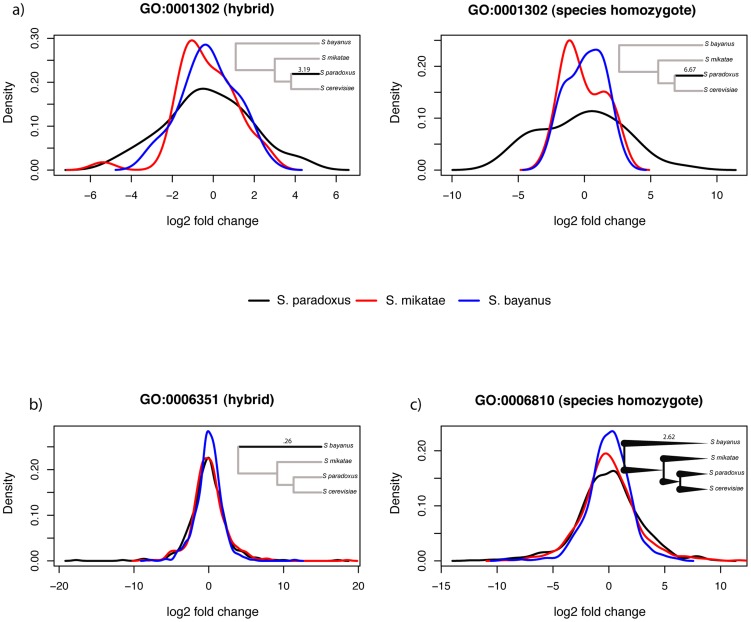
Figure 4. Lineage-specific regulatory evolution and constraint in yeast pathways, inferred from experimental expression measurements.
Each panel shows kernel density estimates of the distributions of experimental gene expression measurements among the genes of one yeast Gene Ontology process term, whose evolutionary history was inferred with strong support. In a given panel, each trace reports the expression levels of the genes of the indicated pathway, from the allele of the indicated yeast species in a hybrid or in the purebred homozygote of a species, normalized with respect to the analogous measurement in S. cerevisiae and with respect to branch length. Inset cartoons represent the model inferred with AIC weight >80% for the indicated pathway (see Tables 1 and 2). (a) Allele-specific expression from measurements in diploid hybrids (left) and total expression measurements in species homozygotes (right) for the 38 genes of GO:0001302, replicative cell aging, supporting a model of accelerated evolution in S. paradoxus; in the inset, the number above the bolded branch reports the inferred shift in the rate of regulatory evolution along that lineage. (b) Allele-specific expression from measurements in diploid hybrids for the 462 genes of GO:0006351, transport, supporting a model of constraint in S. bayanus; in the inset, the number above the bolded branch reports the inferred shift in the rate of regulatory evolution along that lineage. (c) Total expression measured in species homozygotes for the 175 genes of GO:0006281, transcription, supporting an Ornstein-Uhlenbeck model of universal constraint; in the inset, the number above the tree reports the inferred value of the constraint parameter. Note that in (c), the width of the distribution of expression differences between a given species and S. cerevisiae correlates inversely with the sequence divergence of that species, as expected if selective constraint on expression renders the estimate of evolutionary distance from genome sequence an increasing over-estimate of expression change.