Table 1. Top-scoring fitted models of cis-regulatory evolution in yeast pathways from experimental expression measurements.
| GO term | N | Model | wAIC | Constraint or shift parameter |
| 34599 | 57 | Ornstein-Uhlenbeck | 0.899405768 | 49.97745883 |
| 6355 | 433 | S. bayanus shift | 0.837382338 | 0.230918849 |
| 6351 | 462 | S. bayanus shift | 0.849912647 | 0.258701476 |
| 1302 | 38 | S. paradoxus shift | 0.859866949 | 3.197059161 |
| 6897 | 73 | S. paradoxus shift | 0.965743399 | 4.292287639 |
| 6338 | 45 | S. cerevisiae shift | 0.840339574 | 0.037806902 |
| 42254 | 136 | Ornstein-Uhlenbeck | 0.924785133 | 3.733770466 |
| 6364 | 177 | Ornstein-Uhlenbeck | 0.902358815 | 3.079387696 |
| 44255 | 13 | S. paradoxus shift | 0.945799302 | 11.43989834 |
| 54 | 11 | S. paradoxus shift | 0.91523272 | 9.314688245 |
| 16310 | 188 | S. bayanus shift | 0.902247359 | 0.188381056 |
| 8152 | 243 | S. bayanus shift | 0.844716856 | 0.043114988 |
| 6629 | 136 | S. bayanus shift | 0.91650274 | 0.005082617 |
| 122 | 71 | S. bayanus shift | 0.819216472 | 0.040060263 |
| 30437 | 45 | S. paradoxus shift | 0.931136455 | 4.060128813 |
Each row reports the results of phylogenetic inference of the evolutionary history of gene regulation for one yeast Gene Ontology process term, from experimental measurements of cis-regulatory variation in interspecific yeast hybrids. 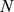 , number of genes in the indicated GO term for which expression measurements were available in all species. Model, best-fit model from among the five possible Brownian motion models of evolutionary rate shift in lineages of the Saccharomyces phylogeny (see Figure 1a), the Ornstein-Uhlenbeck (OU) model of universal constraint, and the equal-rates model involving no lineage-specific differences in evolutionary rate. wAIC, Akaike Information Criterion weight of the indicated model. Constraint or shift parameter, fitted value of the strength of purifying selection or the shift in the rate of regulatory evolution on the indicated lineage, when the best-fit model was the OU model of constraint or a Brownian motion lineage-specific evolutionary rate model, respectively.
, number of genes in the indicated GO term for which expression measurements were available in all species. Model, best-fit model from among the five possible Brownian motion models of evolutionary rate shift in lineages of the Saccharomyces phylogeny (see Figure 1a), the Ornstein-Uhlenbeck (OU) model of universal constraint, and the equal-rates model involving no lineage-specific differences in evolutionary rate. wAIC, Akaike Information Criterion weight of the indicated model. Constraint or shift parameter, fitted value of the strength of purifying selection or the shift in the rate of regulatory evolution on the indicated lineage, when the best-fit model was the OU model of constraint or a Brownian motion lineage-specific evolutionary rate model, respectively.
