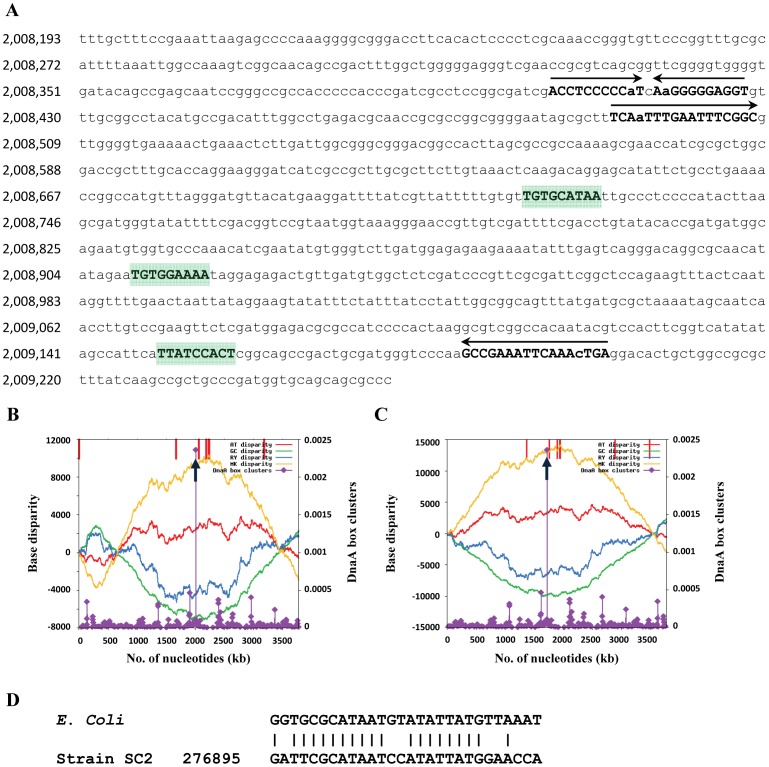
Figure 2. Prediction of the oriC region by Ori-Finder.
(A) 1,063-bp sequence (2,008,193 bp to 2,009,255 bp) of the predicted oriC site. Three dnaA box motifs identified using the Escherichia coli specific dnaA boxes are bold-faced and highlighted. Palindromic repeats identified in this region are marked by arrows at the top. (B, C) The Z-curves measuring the disparity between the percent content of AT (red lines), GC (green lines), RY (blue lines) and MK (yellow lines) for the original sequence (B) and the rotated sequence (C). It should be noted that the coordinate origin of the rotated sequence begins and ends in the maximum of the GC disparity curve. Short vertical red lines at the top show the locations of indicator genes, such as dnaA, dnaN, gidA, and hemE. The upward black arrow indicates the position of the predicted oriC. Purple peaks with diamonds indicate DnaA box clusters. (D) Pairwise alignment between the dif sites located in the genomes of E. coli and strain SC2. In strain SC2, the dif-like sequence is located from nucleotide position 276,895 to 276,922 (almost halfway of the deduced oriC) and matches at 20 nucleotide positions with the 28-bp dif sequence of E. coli.