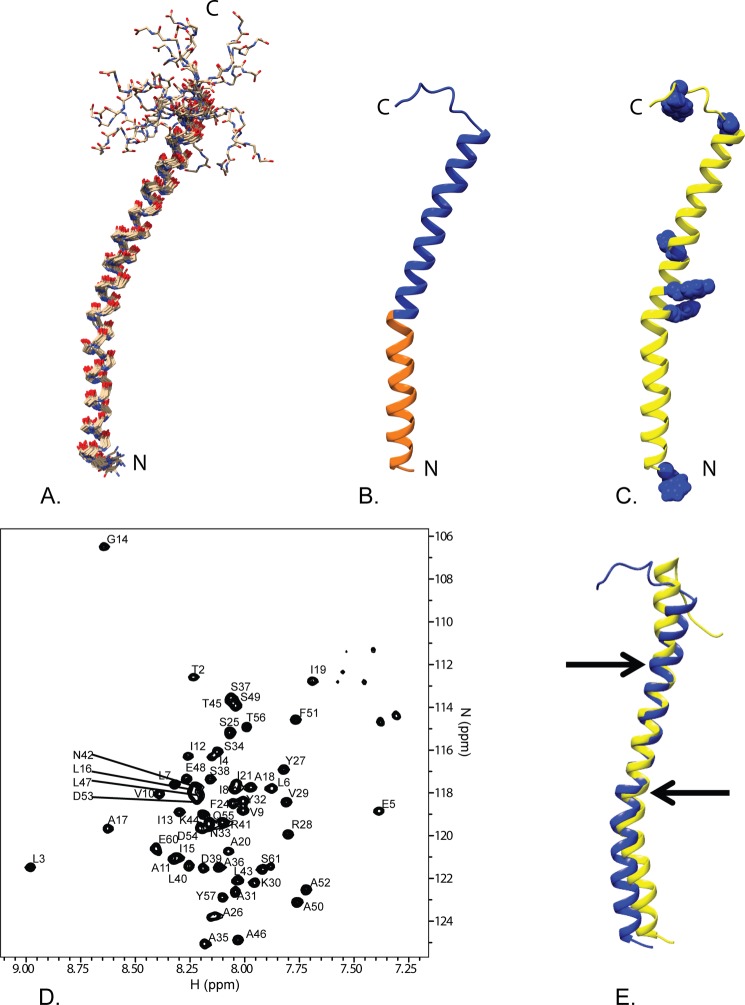
FIGURE 2.
Overview of the solution NMR structure of G. sulfurreducens PilA. A, overlay of the ensemble of 18 structures that did not contain NOE violations >0.5 Å or dihedral angle violations >5°. B, ribbon diagram of the selected conformer (see “Materials and Methods”) with the highly conserved core domain corresponding to amino acids 1–22 colored in orange and the rest of the protein colored blue. C, ribbon diagram of the selected conformer with the aromatic residues shown in blue space filling. The amino terminus and carboxyl terminus are indicated by N and C, respectively. D, 15N TROSY spectrum of GSu PilA with backbone amide assignments. Data were collected at 750 MHz on an Agilent VNMR spectrometer. E, overlay of the homology model of GSu PilA (38) and the experimentally determined GSu PilA structure. The arrows indicate where the degree of bend differs between the structures near residues 22 and 42. To emphasize the differences, the structures were aligned using residues 23–41.