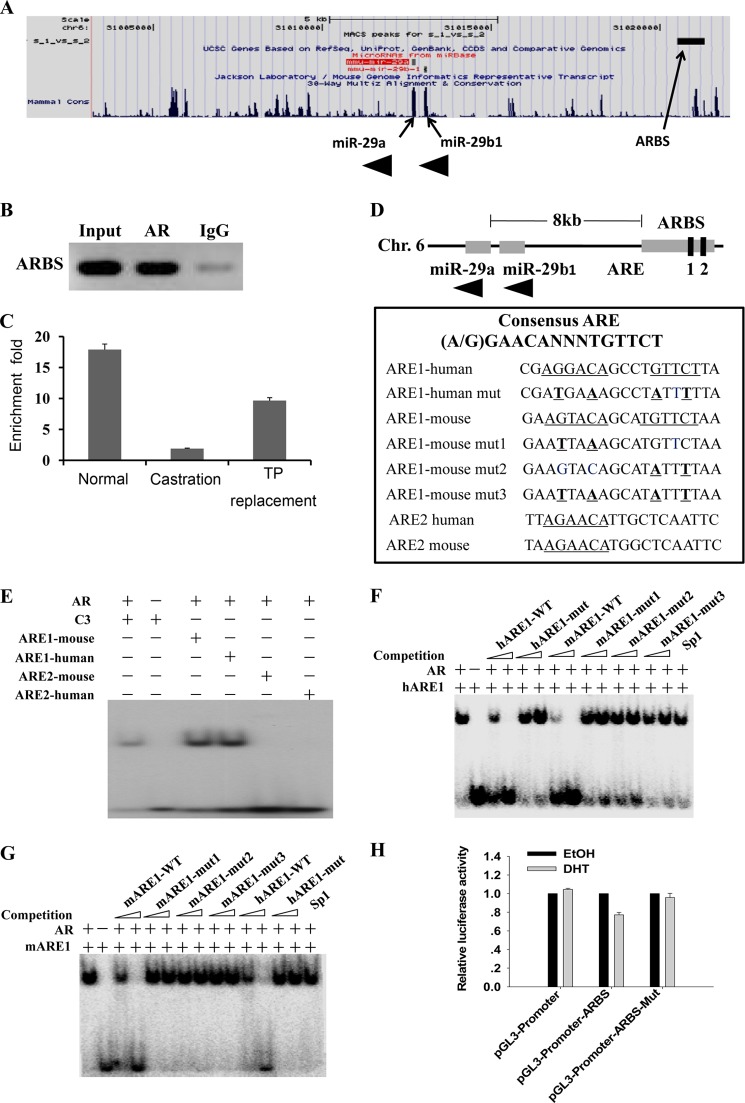
FIGURE 3.
Characterization of AR-binding sites associated with potential AR target gene miR-29b1a. A, screen shots of the UCSC Genome Browser mm9 showing AR-binding sites around miR-29b1a. The figure was generated by uploading one file containing unique ChIP-Seq sites in the BED format to the UCSC Genome Browser. The ChIP-Seq peak is shown at the top, and the AR target genes are shown at the bottom. B, validation of the ARBS associated with miR-29b1a by ChIP-PCR using anti-AR antibody. C, validation of the AR-binding sites associated with miR-29b1a by ChIP-quantitative PCR using anti-AR antibody. Enrichment -fold values were calculated using IgG enrichment as a control (mean ± S.D. (error bars), n = 3). ChIP samples were prepared from epididymidis of normal mice (Nor), mice castrated at 3 days (Cas), and mice castrated at 3 days and supplemented with TP for an additional 2 days. D, the ARBS included two potential conserved AREs; the wild-type and mutated sequences of the two AREs are listed. E–G, AR bound ARE1 but not ARE2 as demonstrated by EMSAs. E, AR bound ARE1 but not ARE2. The ARE of C3 gene served as a positive control. Control incubation performed in the absence of protein is presented in lane 2. In F and G, competitors are indicated above, and amounts of competitors used are given in molar excess. Competitors include unlabeled AREs, mutant AREs, and a consensus DNA-binding site for Sp1. H, ARBS identified by ChIP-Seq functions as an enhancer. The ARBSs with wild-type and mutated ARE1 were subcloned into the pGL3-Promoter plasmid. Plasmids were cotransfected with pcDNA3.1-Ar into HEK 293T cells, and luciferase activities were measured after treatment with ethanol or 100 nm DHT (mean ± S.D. (error bars), n = 3). Chr., chromosome; mARE, mouse ARE; hARE, human ARE; mut, mutant.