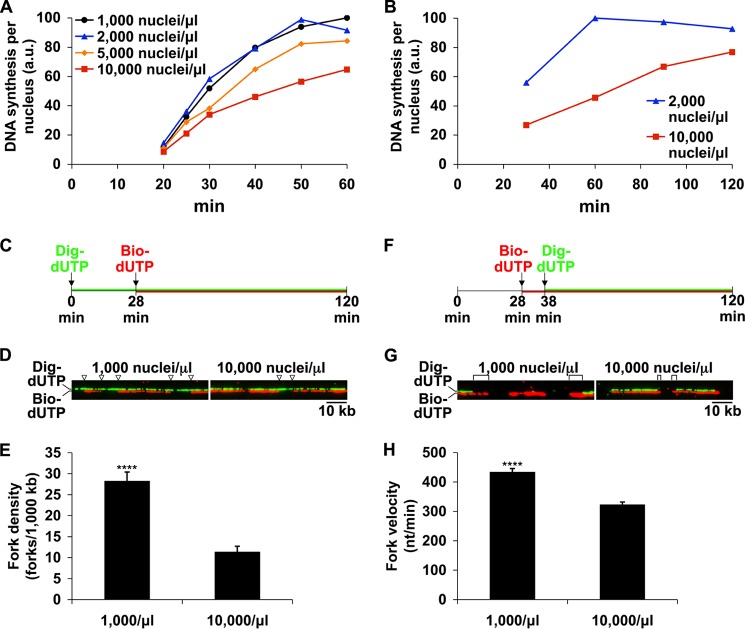
FIGURE 1.
High N/C ratio reduces DNA synthesis by reducing origin firing and slowing replication fork progression. A and B, interphase extracts were supplemented with [α-32P]dATP and either 1,000, 2,000, 5,000, or 10,000 nuclei/μl. Extracts were incubated at 21 °C, and DNA synthesis was monitored at the times indicated. The time scale of 32P incorporation varied somewhat between extracts, so the data shown are from individual experiments and are representative of multiple, independent experiments. C, to analyze replication fork density, bio-dUTP was added at 28 min to mark sites of ongoing replication. Total DNA was labeled with dig-dUTP. D, DNA fibers labeled as in C were prepared and stained as described under “Experimental Procedures.” The locations of replication forks at 28 min on two sample fibers are indicated by arrowheads. The bio-dUTP and dig-dUTP signals were photographed separately and merged at a slight vertical offset to facilitate inspection of each labeling pattern. E, a large number of fibers (at least 15 Mb of DNA) labeled as in C were analyzed for the presence of replication forks. Error bars, S.E. ****, significant at p < 0.0001. F, to determine fork velocity, bio-dUTP was added to extracts during early S phase and followed 10 min later with dig-dUTP to mark the extent of fork progression. G, sample fibers labeled as in F are shown. Digoxigenin-negative tracts used to determine fork progression are marked with brackets. H, fork progression, in nt/min, was determined for at least 260 forks at both low and high N/C ratios. Mean velocity ± S.E. is shown. ****, significant at p < 0.0001 (Student's t test). a.u., arbitrary units.