FIGURE 3.
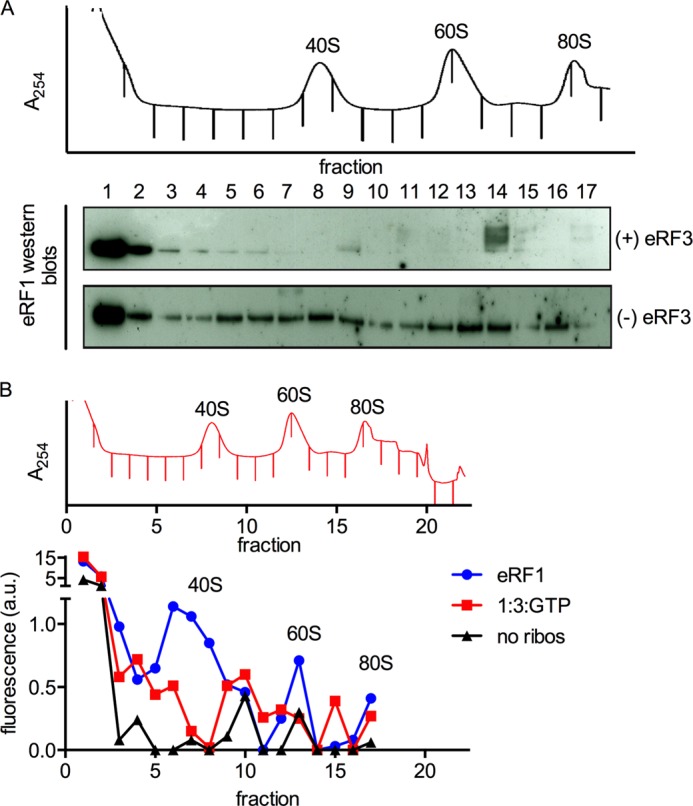
eRF1 remains associated with ribosomal subunits after peptide release and subunit dissociation. A, eRF1 is detected throughout the gradient in the absence of eRF3. Termination complexes (∼70 nm) were reacted with stoichiometric eRF1, with or without eRF3 (1 μm), and separated by sucrose density gradient centrifugation. A trace of absorbance at 254 nm is shown, and the positions of the ribosomal components are labeled. Fractions were collected as indicated by the tick marks in the absorbance trace and analyzed for the presence of eRF1 by Western blotting with standard ECL techniques with an antibody against eRF1. eRF1 is almost exclusively at the top of the gradient in the presence of eRF3 but co-sediments substantially with the 40S, 60S, and 80S peaks in the absence of eRF3. B, eRF1 is detected in the 40S and 60S peaks in the absence of eRF3. Termination complexes were prepared and reacted with eRF1 and/or eRF3, and sucrose density gradient centrifugation was performed as described in A. The results of a quantitative Western blot are shown below the 254 nm absorbance trace. “No ribos“ indicates eRF1 was centrifuged without termination complexes as a negative control. Western blotting was performed using fluorescently labeled secondary antibodies (LICOR). The signal was quantitated using Odyssey and plotted on the y axis in arbitrary units of fluorescence intensity. Note that the y axis is broken to show the fluorescence in fractions 1 and 2.
