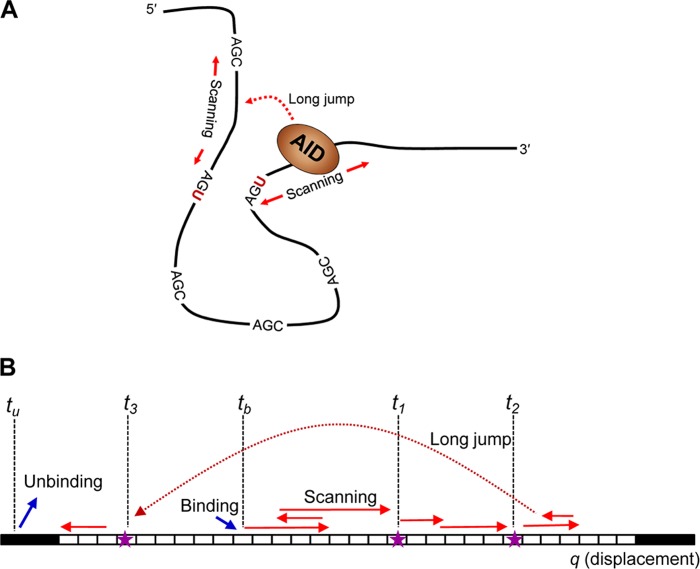
FIGURE 1.
Events leading to a mutation pattern on an ssDNA acted upon by a deaminase enzyme, e.g. AID. A, schematic representation of AID motion (scanning and jumps) and catalysis in three-dimensional space on ssDNA containing multiple AGC hot motifs. B, a projection of AID actions in one-dimensional space for a substrate consisting of a number of trinucleotide motifs (white boxes) that can be deaminated by the enzyme. The enzyme binds to the substrate on a random sequence-independent position at time tb and moves along the substrate by a sequence of random motions including a combination of scanning (sliding or short hops) and possibly long jumps or intersegmental transfers. The enzyme catalyzes C→U mutations along its trajectory, dropping markers (stars) at time t1, t2, … The enzyme eventually unbinds at time tu. This sequence of events can be mapped onto an abstract one-dimensional sequence space on which the stochastic model is formulated.