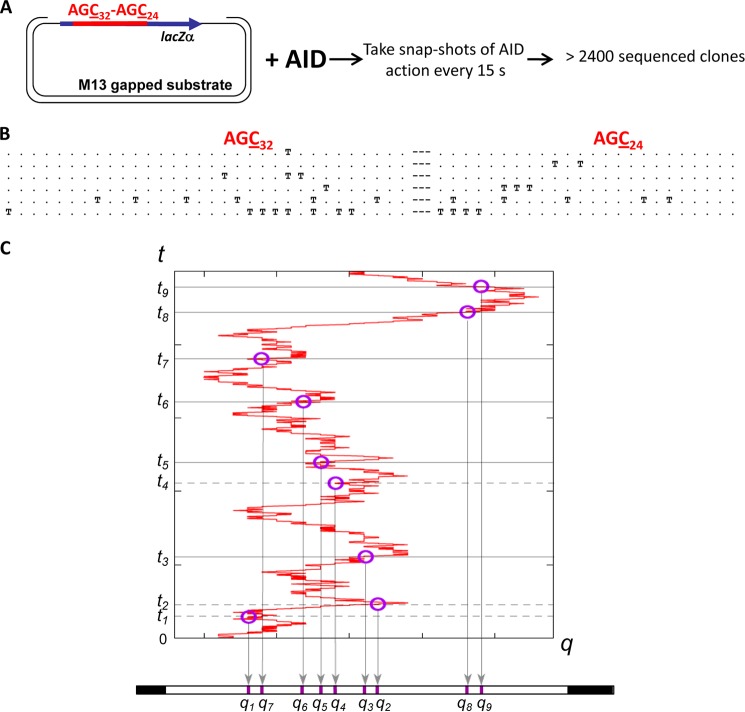
FIGURE 2.
Time-course experiment to analyze AID random scanning and stochastic deamination on ssDNA. A, an M13mp2 phage construct containing an ssDNA region with 32 AGC deamination motifs on the left side and 24 AGC motifs on the right side separated by a 9-nt linker region was subjected to AID deamination for a duration from 15 s to 10 min at 37 °C. AID deaminations at AGC motifs are detected as C→T mutations in individual M13 mutant phage clones (25). B, representative mutated clones with 1, 2, 3, 4, and ≥5 mutations. T and dot symbols denote deaminated and non-deaminated AGC motifs, respectively. C, a scanning trajectory of AID on an ssDNA combined with stochastic deamination events depositing a random sequence of markers at positions q1, q2, … at time t1, t2, … on the ssDNA substrate.