FIGURE 1.
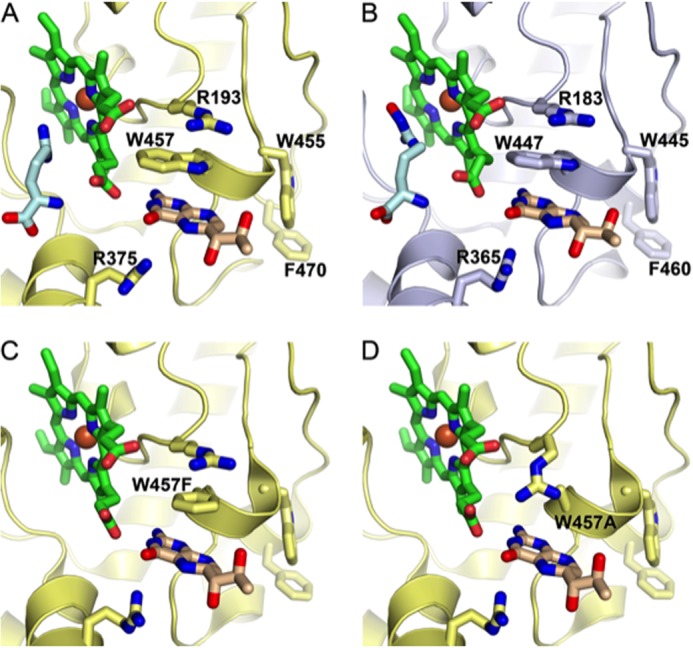
Human eNOS and murine iNOS have closely related structures. A, the active site of iNOS (yellow), showing the heme group (green) and arginine substrate (pale blue). B, the active site of eNOS (gray), demonstrating its close structural similarity with iNOS. Structural superposition of human eNOS and murine iNOS (PDB codes 3NOS and 1NOD, respectively) was performed using PDBe Fold. The top-ranked root mean square deviation between 365 matched residues in a single protein chain was 0.81Å. C, the active site of iNOS showing the W457F mutation. D, the active site of iNOS showing the W457A mutation. Figures were produced using PyMOL on the basis of Fig. 1 by Wang et al. (15) using PDB codes 3NOS (eNOS), 1NOD (iNOS), 1JWJ (iNOS W457F), and 1JWK (iNOS W457A).
