FIGURE 1.
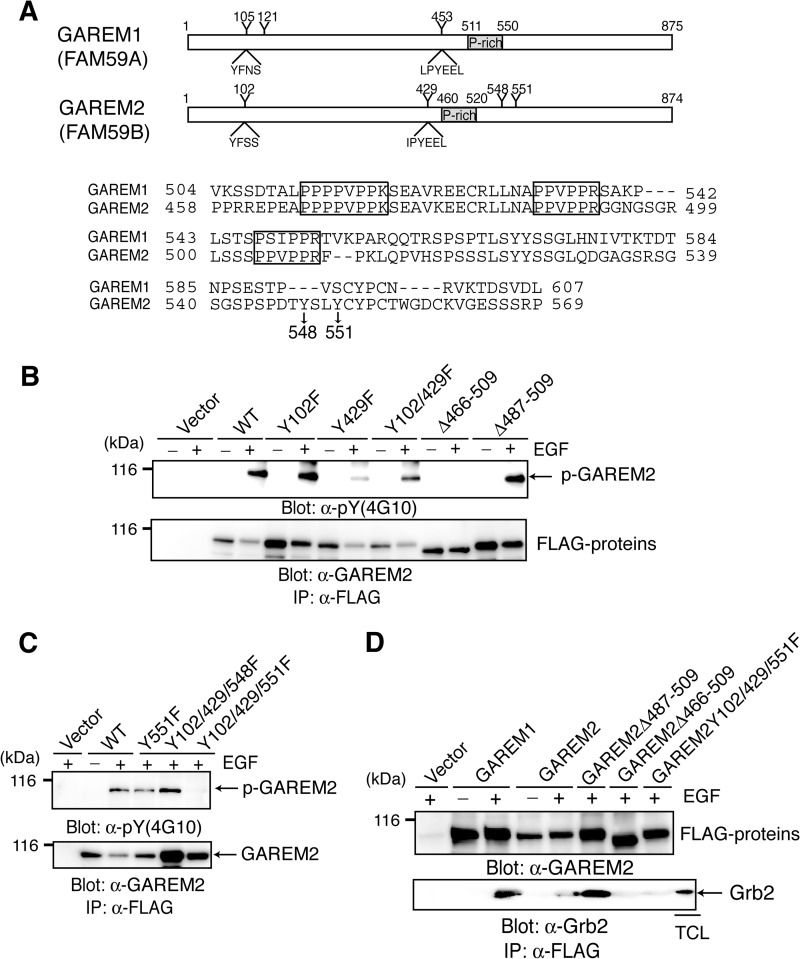
GAREM2, like GAREM1, is a Grb2-binding protein. A, schematics of the primary structure of GAREM1 and GAREM2 (upper panel). Representative tyrosine residues and the surrounding amino acid sequence in GAREM1 and GAREM2 are indicated. Numbers indicate amino acid residues. Also shown are amino acid sequences of the proline-rich region of GAREM1 and GAREM2 (lower panel). Proline-rich (P-rich) regions that may bind the SH3 domain are indicated in the box. Tyrosines 548 and 551 in GAREM2, which are not conserved in GAREM1, are indicated by arrows. B and C, the unique tyrosine phosphorylation site (Tyr-551) in GAREM2. COS7 cells were transfected with plasmids encoding the FLAG-tagged construct of GAREM2 derivatives in which tyrosine residues (Y) were substituted with phenylalanine (F) and deleted (Δ) in the proline-rich motifs. The number of substituted or deleted amino acid residues is indicated. Immunoprecipitation (IP) studies were performed with lysates from COS-7 cells transfected with the empty vector or indicated plasmids. Cells were treated with (+) or without (−) EGF stimulation for 10 min, and each FLAG-tagged molecule was immunoprecipitated with the corresponding anti-FLAG antibody. Immunoblotting was performed using anti-phosphotyrosine (top panel) and anti-FLAG antibodies (bottom panel). D, the proline-rich motifs in GAREM2 enable binding to Grb2. This binding is dependent on tyrosine phosphorylation of GAREM upon EGF stimulation. Coimmunoprecipitation studies were performed with the anti-FLAG antibody by using the lysates of COS-7 cells transfected with plasmids carrying FLAG-GAREM2 derivatives and treated with EGF. Immunoblotting was performed with anti-FLAG (top panel) or anti-Grb2 antibodies (bottom panel).