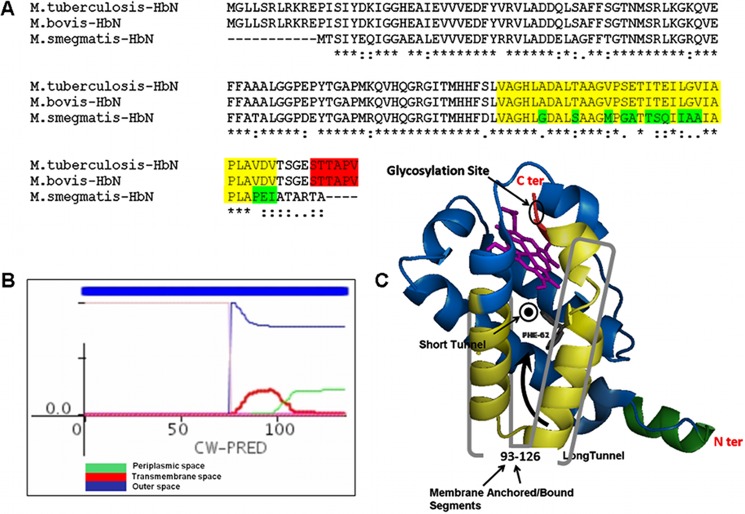
FIGURE 5.
In silico analysis of the primary sequence of HbN of Mtb. A, multiple-sequence alignment of the amino acid sequence of HbN from M. tuberculosis, M. bovis, and M. smegmatis. The segment shown in fluorescent yellow depicts extracellular and membrane-anchored/bound regions predicted in the HbN of M. tuberculosis and M. bovis. The glycosylation site in HbN is highlighted in red. The differences within the membrane-anchored/bound region in the HbN of M. smegmatis are highlighted in green. B, prediction of membrane-bound/anchored region in Mtb HbN using CW-PRED that assigns a score of 1.0 to such proteins. The output here depicts three different colors to define the triplet state in which a residue can be present, namely periplasmic, extracellular, or transmembrane. Predictions for all of the states are combined in the output, and the analysis indicated that the region spanning amino acid residues 93–126 at the C terminus may constitute a potential extracellular association of MtbRa-HbN. C, a hypothetical schematic model of HbN depicting the site of glycosylation and the potential membrane-anchored segment shown in yellowish green.