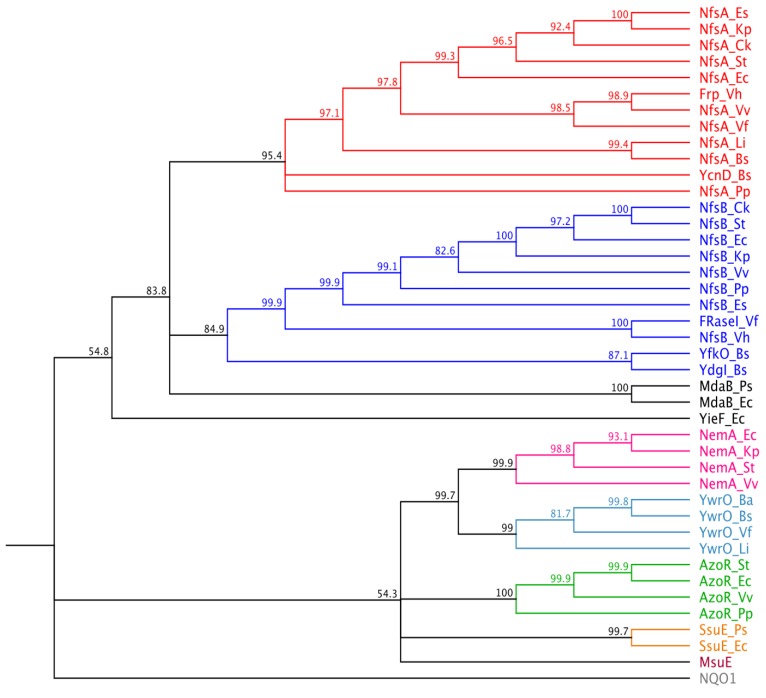
Figure 2.
Phylogenetic analysis of MsuE and other experimentally validated bacterial nitroreductase families. MsuE (dark red) and two SsuE orthologues from E. coli and Pseudomonas syringae pv. tomato DC3000 (orange) were genetically aligned against members of the NfsA (red), NfsB (dark blue), MdaB (black), YieF (black), NemA (pink), YwrO (light blue) and AzoR (green) nitroreductase families, and a human NQO1 out-group (grey), using the online tool ClustalW2. With the exception of the msuE and ssuE genes, nitroreductase sequences are as previously presented by Prosser et al. [14]. Following the nomenclature of that paper, the two letter suffix following the enzyme name indicates the bacterial strain of origin: Ba, B. amyloliquefaciens; Bs, Bacillus subtilis; Ck, Citrobacter koseri; Es, Enterobacter sakazakii; Ec, Escherichia coli; Kp, Klebsiella pneumonia; Li, Listeria innocua; Pa, Pseudomonas aeruginosa; Pp, Pseudomonas putida; Ps, Pseudomonas syringae; St, Salmonella typhi; Vf, Vibrio fischeri; Vh, Vibrio harveyi; Vv, Vibrio vulnificus. The phylogenetic tree was constructed using the online tool MrBayes 3.2.1 [27] and bootstrap confidence values are indicated at major branchpoints. The final figure was generated using Geneious version 6.1 [28].