Table I.
Data-Collection, Refinement and Validation Statistics of the Structure of the P32G β2m•Nb24 Complex
| Data Set | P32G β2m•Nb24 |
|---|---|
| Data-collection | |
| X-ray source | Diamond IO3 |
| X-ray wavelength (Å) | 0.97950 |
| Temperature (K) | 100 |
| Space group | P 42212 |
| Unit-cell parameters | |
| a, b, c (Å) | 96.8, 96.8, 167.8 |
| α, β, γ(°) | 90.0, 90.0, 90.0 |
| Resolution range (Å) | 48.43–2.6 (2.67–2.60) |
| Total/Unique reflections | 362392/25281 |
| Rmerge (%)a | 35.7 (392) |
| Rmeas (%)b | 37.0 (406) |
| Data completeness (%) | 99.9 (99.8) |
| Average I/σ | 8.5 (1.0) |
| Redundancy | 14.3 (14.7) |
| Wilson B factor (Å2) | 56.1 |
| CC(1/2) | 99.7 (62.3) |
| Refinement | |
| Correlation coefficients | |
| Correlation coefficient Fo − Fc | 0.948 |
| Correlation coefficient Fo − Fc Free | 0.914 |
| Rwork/Rfreec | 22.84/27.69 |
| Total number | |
| Amino acid residues | 445 |
| Water molecules | 50 |
| Ligand atoms | 8 |
| rmsd | |
| Bond length (Å) | 0.0150 |
| Bond angles (°) | 1.5567 |
| Average atomic B-factor (Å2) | |
| Protein atoms | 58.67 |
| Solvent atoms | 35.0 |
| Ramachandran plot (%) | |
| Favored regions | 97.70 |
| Allowed regions | 2.30 |
| Disallowed regions | 0 |
| PDB entry | 4KDT |
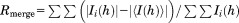
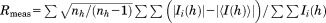
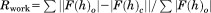 , F(h)o and F(h)c are observed and calculated structure factor amplitudes, respectively. A random subset of data (5%) was used for the Rfree calculation.
, F(h)o and F(h)c are observed and calculated structure factor amplitudes, respectively. A random subset of data (5%) was used for the Rfree calculation.
