Figure 1.
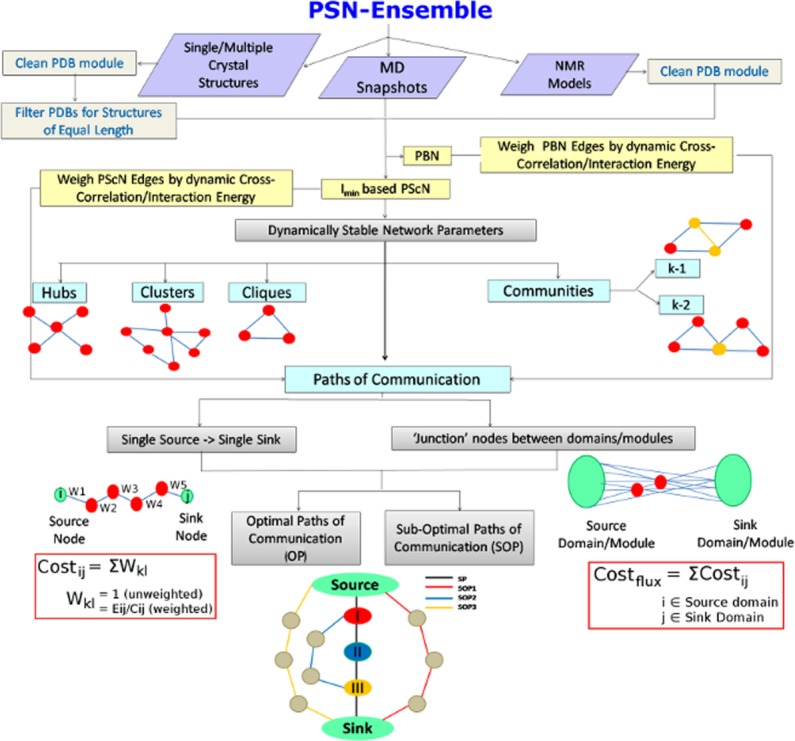
A flowchart representing the workflow in PSN-Ensemble. The package is compatible with crystal structure (single or multiple structures), MD simulation, and NMR ensemble structures. Various network parameters for characterizing the topological organization in proteins [hubs, clusters, cliques, communities (k-1/k-2)] can be obtained at a given Imin (see Methods section) from PScN. The network parameters to characterize long-range communication in terms of paths and cost of communication [OPs and SOPs] can be calculated based on PScN or PBN. A schematic description of each of the network parameters is also included in the flowchart for easy comprehension. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
