Fig. 1.
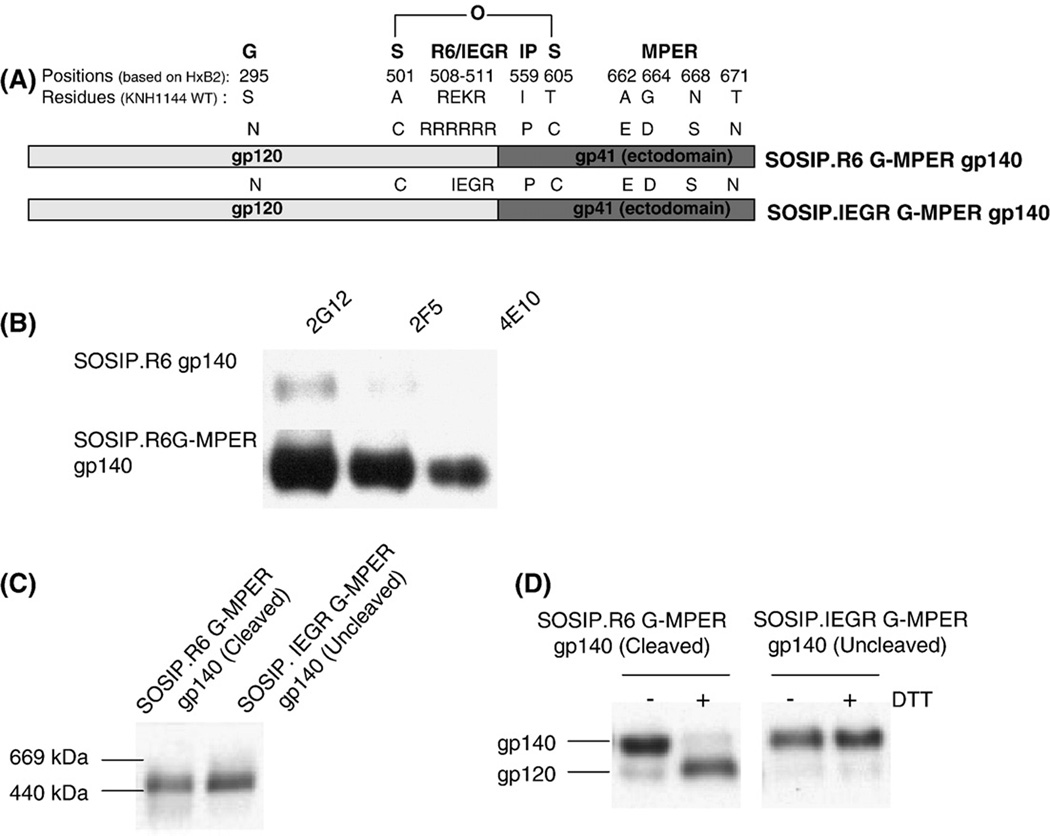
Expression and analysis of soluble, cleaved (SOSIP.R6 G-MPER) and uncleaved (SOSIP.IEGR G-MPER) gp140s from HIV-1 KNH1144. (A) Schematic representation of the various mutations made in the KNH1144 primary sequence to generate a stabilized soluble trimeric gp140 (SOS, IP), to promote (R6) or disrupt (IEGR) cleavage, to introduce the 2G12 epitope (S295N), and to improve 2F5 and 4E10 binding (MPER - A662E, G664D, N668S and T671N). (B) The epitopes for MAbs 2G12, 2F5 and 4E10 were introduced into the KNH1144 SOSIP.R6 G-MPER gp140 sequence by site-directed mutagenesis, to create the SOSIP.R6 G-MPER variant. Binding of these MAbs to the two gp140s was determined by immunoprecipitation. (C) Unpurified cell culture supernatants containing the same cleaved or uncleaved gp140s were analyzed by BN-PAGE and western blotting with MAb CA13 (ARP3119) that binds to the C1 region of gp120. The migration positions of relevant molecular weight standards are marked on the left of the gel. Both the cleaved and uncleaved gp140s migrate as trimers. (D) The same cleaved (lanes 1 and 2) and uncleaved (lanes 3 and 4) gp140s were analyzed by SDS-PAGE in the presence or absence of the reducing agent, DTT, as indicated, followed by western blotting. The SOSIP.R6 G-MPER gp140 is fully cleaved and hence dissociates to gp120 in the presence of DTT (lane 2), whereas the SOSIP.IEGR G-MPER protein is not cleaved (lane 4).
