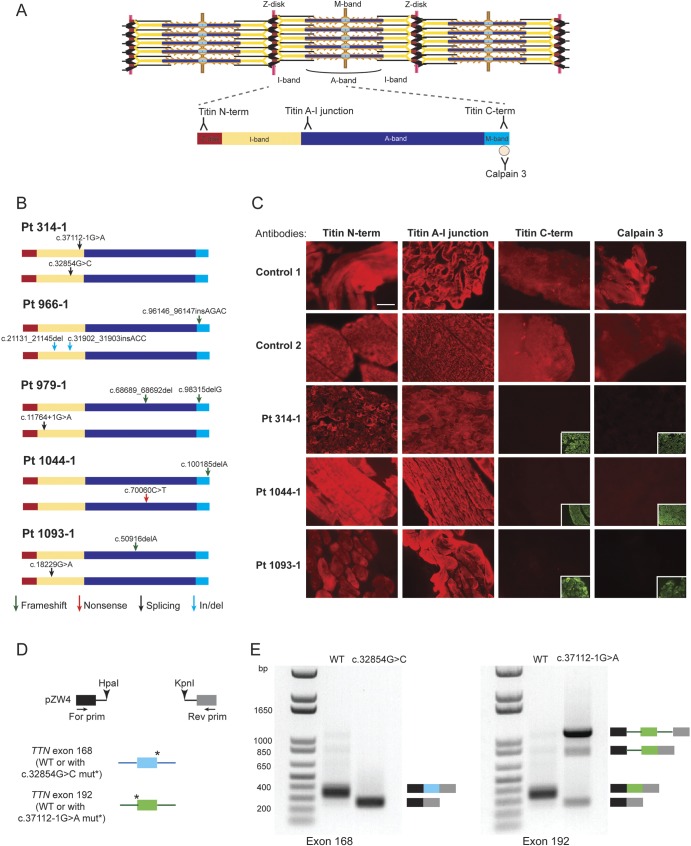
Figure 2. TTN mutations associated with centronuclear myopathy cause a loss of titin C-terminal region in patient muscles.
(A) Locations of the antibody epitopes used in immunofluorescence analysis are depicted on the skeletal muscle titin isoform N2A (NM_133378). The Z-disk (red), I-band (yellow), A-band (dark blue), and M-band (light blue) regions of a titin molecule spanning halfway of a sarcomere are represented. (B) Locations of the TTN mutations in each patient are represented on the N2A isoform. Arrows depict the type of mutations: frameshift (green), nonsense (red), splicing (black), in-frame insertion/deletion (blue). (C) Immunofluorescence analysis of patient and age-matched healthy control muscles. Frozen muscle sections were stained with titin N-terminal, A–I junction, C-terminal, or calpain 3 antibody (red); and with anti-α-actinin antibody (green). Staining with the control anti-α-actinin antibodies are presented in the inset lower right panels where the titin C-terminal or calpain 3 antibodies showed no signal. (D) Overview of the hybrid minigene splicing assay. Wild-type exon 168 and 192 (numbering based on N2A isoform) were cloned with their flanking introns into the pZW4 splicing reporter construct between HpaI and KpnI restriction sites. The mutations at the donor or acceptor splice sites were introduced by site-directed mutagenesis. Wild-type or mutant hybrid minigene containing plasmids were transfected into HEK293 cells, followed by RNA extraction and reverse transcriptase–PCR using primers flanking the hybrid constructs. (E) Results of the hybrid minigene splicing assay. The minigenes containing wild-type exon 168 or exon 192 were spliced correctly. The c.32854G>C mutation at the donor splice site of exon 168 caused exon skipping (left panel). The c.37112-1G>A mutation at the acceptor splice site of exon 192 resulted in the majority of transcripts to remain unspliced, as well as exon skipping or intron retention in a subset of transcripts (right panel). Scale bar = 40 μm. For = forward; mut = mutation; Pt = patient; Rev = reverse; WT = wild-type.