Table 1. Naturally occurring and synthetic nucleosome-positioning or nucleosome-repelling sequences used in this study.
| Sequence Name | Species | Genomic Position | Nucleosome Affinity |
| poly(dA:dT) (“dA”) | Budding yeast (S. cerevisiae) | Chr III, 38745 – 39785 bp (Ref. [23]) | ∼3-fold in vivo nucleosome depletion relative to average genomic DNA (Fig. 2E of Ref. [22]);  increase in energy of nucleosome formation in vitro relative to 5S (Fig. 8D of Ref. [22]) (estimates based on similar sequences) increase in energy of nucleosome formation in vitro relative to 5S (Fig. 8D of Ref. [22]) (estimates based on similar sequences)
|
| GC-rich (“CG”) | Human | Chr Y, 4482107 – 4481956 bp (Ref. [22]) | (not determined) |
| 5S | Sea urchin (L. variegatus) | 20 bp–165 bp from the Mbo II fragment containing 5S rRNA gene (Ref. [45]) |
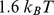 decrease in energy of nucleosome formation compared to E8 in vitro (Ref. [8]) decrease in energy of nucleosome formation compared to E8 in vitro (Ref. [8]) |
| 601TA (“TA”) | synthetic, strong nucleosome positioning sequence (Refs. [8], [59], [60]) | N/A |
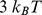 decrease in energy of nucleosome formation compared to E8 in vitro (Ref. [8]) decrease in energy of nucleosome formation compared to E8 in vitro (Ref. [8]) |
| E8 | synthetic random (Refs. [8], [60]) | N/A | (used as a reference) |
The sequences described here were chosen because each has been found to have significant effects on in vivo nucleosome positions and/or in vitro nucleosome affinities, as shown in the rightmost column. The exception is the GC-rich sequence from humans: although its nucleosome affinity has not been directly determined either in vivo or in vitro, it is predicted to correlate with high nucleosome occupancy because of its high G+C content [17] and is occupied by a nucleosome(s) in vivo according to micrococcal nuclease digestion [22]. Two-letter abbreviations given in parentheses under each full sequence name will be used in figure legends in the rest of this work.
