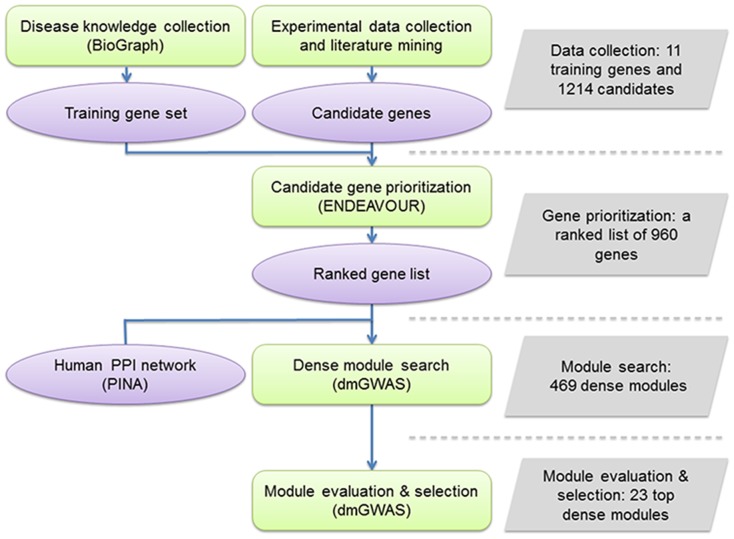
Figure 1. The workflow of this study.
First, the biological knowledgebase BioGraph [34] was explored to identify training gene set, and candidate genes were collected from previous studies and publications. We obtained 11 training genes and 1214 candidate genes in this data collection step. Second, a computational method ENDEAVOUR [13] was utilized to prioritize the candidate genes. In this step, a ranked list of 960 candidate genes that could be recognized by ENDEAVOUR was generated. Third, dmGWAS [33] was employed to search for the dense modules upon human protein-protein interaction (PPI) network collected by Protein Interaction Network Analysis (PINA) platform [53]. This resulted in 469 dense modules. Finally, the 469 modules were evaluated and the top 23 ones were selected as promising modules.