Abstract
Shipworms are marine bivalve mollusks (Family Teredinidae) that use wood for shelter and food. They harbor a group of closely related, yet phylogenetically distinct, bacterial endosymbionts in bacteriocytes located in the gills. This endosymbiotic community is believed to support the host's nutrition in multiple ways, through the production of cellulolytic enzymes and the fixation of nitrogen. The genome of the shipworm endosymbiont Teredinibacter turnerae T7901 was recently sequenced and in addition to the potential for cellulolytic enzymes and diazotrophy, the genome also revealed a rich potential for secondary metabolites. With nine distinct biosynthetic gene clusters, nearly 7% of the genome is dedicated to secondary metabolites. Bioinformatic analyses predict that one of the gene clusters is responsible for the production of a catecholate siderophore. Here we describe this gene cluster in detail and present the siderophore product from this cluster. Genes similar to the entCEBA genes of enterobactin biosynthesis involved in the production and activation of dihydroxybenzoic acid (DHB) are present in this cluster, as well as a two-module non-ribosomal peptide synthetase (NRPS). A novel triscatecholate siderophore, turnerbactin, was isolated from the supernatant of iron-limited T. turnerae T7901 cultures. Turnerbactin is a trimer of N-(2,3-DHB)-L-Orn-L-Ser with the three monomeric units linked by Ser ester linkages. A monomer, dimer, dehydrated dimer, and dehydrated trimer of 2,3-DHB-L-Orn-L-Ser were also found in the supernatant. A link between the gene cluster and siderophore product was made by constructing a NRPS mutant, TtAH03. Siderophores could not be detected in cultures of TtAH03 by HPLC analysis and Fe-binding activity of culture supernatant was significantly reduced. Regulation of the pathway by iron is supported by identification of putative Fur box sequences and observation of increased Fe-binding activity under iron restriction. Evidence of a turnerbactin fragment was found in shipworm extracts, suggesting the production of turnerbactin in the symbiosis.
Introduction
Iron is required by all but a few organisms and serves as an important cofactor for many essential enzymes. The insolubility of Fe(III) at physiological pH and aerobic conditions severely limits its bioavailability. In the world's oceans, 99% of the Fe(III) is bound by uncharacterized organic ligands [1], [2]. Similarly, iron is also a limiting nutrient for bacteria in animals, where it is often tightly bound to proteins such as ferritin, transferrin, lactoferrin, or incorporated into heme containing proteins [3], [4]. The ability of a bacterium to acquire iron may be critical to the successful colonization of its host. In response to iron limitation, many bacteria and some fungi produce siderophores, low-molecular weight compounds with a high affinity for Fe(III). Hundreds of siderophores have been characterized, the majority being from terrestrial organisms [5], [6], [7]. Siderophores can be classified by their mode of biosynthesis, non-ribosomal peptide synthetase (NRPS)-dependent or NRPS-independent biosynthesis [8], as well as by their major type of functional binding groups, catechols, hydroxamic acids, and α-hydroxycarboxylic acids.
The class of siderophores that possess the highest stability constants measured to date are the triscatecholate siderophores, including enterobactin [9], [10], a cyclic trimeric ester of 2,3-dihydroxybenzoate (DHB)-L-Ser (Kf = 1049, [11]) and bacillibactin [12], a cyclic trimeric ester of 2,3-DHB-Gly-L-Thr (Kf = 1047.6, [13]). Other examples of cyclic triscatecholate siderophores include salmochelin, a glucosylated derivative of enterobactin [14], cyclic trichrysobactin [15], and streptobactin [16]. The recently reported cyclic trichrysobactin is a trimeric ester of the previously reported chrysobactin, 2,3-DHB-D-Lys-L-Ser [17], and was isolated from the plant pathogen Dickeya chrysanthemi. Streptobactin, a cyclic trimeric ester of 2,3-DHB-L-Arg-L-Thr, was recently reported from the marine-derived actinomycete Streptomyces sp. YM5-799. In addition to the cyclic triscatecholate siderophores, a linear (non-cyclized) triscatecholate siderophore, trivanchrobactin, was reported from the marine isolate Vibrio campbelli DS40M4 [18], [19]. Trivanchrobactin is a linear trimeric ester of the previously reported vanchrobactin, 2,3-DHB-D-Arg-L-Ser [20].
The shipworm endosymbiont Teredinibacter turnerae has been found in numerous genera and species of teredinid bivalves from across the globe [21]. Shipworm endosymbionts have been shown to supplement the nitrogen-poor wood diet by providing a source of fixed nitrogen and are thought to produce cellulolytic enzymes that may assist the shipworm host in the degradation of wood [21]. The genome of T. turnerae T7901 was recently sequenced [22]. Although T. turnerae is an intracellular symbiont, its genome does not show the typical modifications experienced by obligate endosymbionts, most notably reduced genome size, higher A+T content, and loss of DNA repair and transcriptional regulatory genes [23]. Its genome size is 5.1 Mb, with a 50.8% G+C content, and includes genes involved in almost all core metabolic functions, including DNA repair [22]. These observations suggest either T. turnerae is a facultative symbiont, or is part of a recently established symbiosis. To date, T. turnerae has not been detected outside of the shipworm host. Analysis of T. turnerae's genome also reveals nine gene clusters predicted to code for secondary metabolites, constituting nearly 7% of its genome. One of the nine gene clusters was recently reported to synthesize an antibacterial compound in the tartrolon family [24].
Here we report on another secondary metabolite gene cluster from T. turnerae T7901, a cluster that was previously proposed to encode a siderophore biosynthesis gene cluster [22]. This work presents the structural characterization of a novel triscatecholate siderophore, N-(2,3-DHB)-L-Orn-L-Ser, named turnerbactin. The gene cluster responsible for the biosynthesis of turnerbactin is analyzed and a model for its biosynthesis is proposed. The biosynthetic genes involved in turnerbactin production are herein referred to as tnb, for turnerbactin biosynthesis. Turnerbactin represents the first described siderophore from a shipworm symbiont.
Methods
Strain information
Teredinibacter turnerae T7901 was originally isolated from the shipworm Bankia gouldi collected in the wild near Duke University Marine Lab, Beaufort, NC [21].
Culture conditions
T. turnerae T7901 was grown in a low-iron, modified version of shipworm basal medium (SBM) [21], [25] containing 750 ml artificial seawater [26], 250 ml distilled water, 0.1 mM KH2PO4, 0.094 mM Na2CO3, 0.01 mM Na2MoO4•2H2O, 0.5% (w/v) sucrose, 5 mM NH4Cl, 20 mM HEPES buffer (pH 8.0), 0.1 μM EDTA-chelated ferric iron (Sigma), and 1 ml A5+Co trace metal mix [27]. Cultures were grown in 2.8 L Fernbach flasks with 2 L culture medium at 30°C on an orbital shaker (110 rpm). A pre-culture of T. turnerae was prepared by inoculating an iron-replete (10 μM EDTA-chelated ferric iron) SBM liquid culture with a single colony from an iron-replete SBM agar plate. The 2 L cultures were inoculated with 1 mL of overnight-grown pre-culture. Cultures were grown for 2 days, when Fe(III)-binding activity of culture supernatants reached maximum activity as measured by the chrome azurol sulfonate (CAS) assay [28].
To determine CAS activity of iron-replete and iron-limited cultures, duplicate 50 mL SBM cultures of T. turnerae T7901 were grown in 100 mL flasks with the same conditions described above, containing either 10 μM EDTA-chelated ferric iron (iron-replete) or 0.1 μM EDTA-chelated ferric iron (iron-limited). After growth for approximately 38 hours at which point cultures were at late log phase, 1 mL aliquots of all cultures were collected and centrifuged at 10000 rpm for 3 min. Supernatants were tested for CAS activity. Uninoculated iron-replete media showed no CAS activity, indicating that the EDTA from the iron source did not interfere with the assay.
All growth and CAS activity measurements were recorded on a SpectraMax M2 Multidetection Reader (Molecular Devices) with absorbance measured at 600 nm and 630 nm, respectively. CAS activity was calculated as (1-(Absample/Abblank) ×100.
Compound isolation
Culture supernatant was cleared of cells and debris by centrifugation at 10000 rpm for 25 min. Decanted supernatant was incubated with ∼20 g/L Dianion HP20 resin in 2.8 L Fernbach flasks at 4°C on an orbital shaker (110 rpm) for 4 hrs. The resin was collected and washed sequentially with MilliQ water followed by 25%, 50%, and finally 100% isopropanol (IPA). The siderophores eluted with the 25% IPA fraction as detected by the CAS assay. The 25% IPA fraction was concentrated by rotary evaporation in vacuo. The siderophores were purified by reverse-phase high–performance liquid chromatography (RP-HPLC) on a semi-preparative C18 column (10 mm internal diameter ×25 cm length, 5 μm particle size, Ascentis) using water and acetonitrile (ACN) as solvents at a flow-rate of 3 mL/min at 25°C. Both solvents contained 0.05% trifluoroacetic acid (TFA). The mobile phases consisted of a gradient of 10% ACN in water to 15% ACN from 0–10 min, followed by an isocratic step of 15% ACN from 10–15 min, then a gradient from 15% to 25% ACN from 15–35 min, then a final gradient from 25% to 100% ACN from 35–40 min. The eluent was continuously monitored at 215 nm and 320 nm. Fractions were collected manually and immediately tested for CAS activity and then concentrated under vacuum. Collected and pooled fractions were further purified on an analytical RP-Amide-C16 column (4.6 mm internal diameter ×25 cm length, 5 μm particle size, Ascentis) using the same method as above at a flow-rate of 1 mL/min. Pure siderophores were lyophilized. The approximate yields of siderophores from a 4 L culture are 1.2 mg DHB-Orn-Ser, 2.1 mg (DHB-Orn-Ser)2, 2.7 mg turnerbactin, 2.4 mg dehydro-(DHB-Orn-Ser)2, and 1.2 mg dehydro-(DHB-Orn-Ser)3.
The triscatecholate siderophores that have been reported in the literature have not contained dehydrated amino acid constituents. Additionally, with the exception of linear trivanchrobactin, the triscatecholate siderophores have been found in the cyclic form. A control experiment was carried out to rule out the possibility that the dehydrated siderophores and the lack of a cyclic trimer were the result of the purification method. A side-by-side purification of the structurally similar siderophore cyclic trichrysobactin [15] was carried out with siderophores from T. turnerae T7901. Cultures of Dickeya chrysanthemi were grown and siderophores were purified as described by Sandy and Butler [15]. Siderophores from culture extracts of T. turnerae T7901 were purified concurrently and in the same manner. The identity of siderophores from both cultures was confirmed by HPLC retention time, high-resolution electrospray ionization mass spectrometry (HRESIMS), and 1H-NMR.
Mass spectra were obtained on a ThermoElectron LTQ-Orbitrap high-resolution mass spectrometer. Samples were dissolved in 50% methanol and MS analysis was performed in positive mode using electrospray ionization (ESI). All 1-D and 2-D NMR experiments were carried out on a Bruker Avance II Ultrashield Plus 800 MHz instrument with a cryoprobe in d 4-methanol (CD3OD, Cambridge Isotope Laboratories).
UV-visible absorption spectrum of purified turnerbactin in water was collected on a Varian Cary 50 spectrophometer.
Amino acid analysis
A dried preparation of purified turnerbactin (∼1 mg) was hydrolyzed in 200 μL 6 M HCl for 17 hours at 110°C. The solution was brought to room temperature and evaporated to dryness. The dried, hydrolyzed sample was redissolved in 100 μL H2O, to which 200 μL of a 1% (w:v) solution of Marfey's reagent (1-fluoro-2–4-dinitrophenyl-5-L-alanine amide [FDAA]) [29] in acetone and 40 μL 1 M NaHCO3 were added to derivatize the primary amines of the amino acids. The reaction was heated for 1 hour at 40°C, after which 20 μL 2 M HCl was added to stop the reaction. The derivatized sample was analyzed by HPLC on an analytical YMC ODS-AQ C18 column (4.6 mm, i.d. ×250 mm L, Waters Corp.) using a linear gradient from 90% water with 0.1% TFA/10% CH3CN to 60% water with 0.1% TFA/40% CH3CN over 45 min. The eluent was continuously monitored on a Waters UV-visible detector at 340 nm. The derivatized sample was compared to chiral amino acid standards prepared the same way. Peaks were collected and verified by MS.
Chemical extraction of shipworms
A laboratory culture of Lyrodus pedicellatus maintained at the Ocean Genome Legacy was sent to Oregon Health & Science University and stored at room temperature in an aerated aquarium until further processed. L. pedicellatus specimens were removed from wood using pliers, slowly peeling away layers of wood until shipworms were exposed, careful to not damage the shipworms. Six L. pedicellatus shipworms were removed from the wood, rinsed in filter-seawater, pooled, and lyophilized. The resulting dry weight was 85.4 mg. The tissue was then homogenized with a plastic pestle. The homogenized tissue was extracted three times with three volumes of methanol on a rotary mixer for one hour. The methanolic extract was separated from the tissue by centrifugation at 15,000 rpm for 3 min and subsequently dried. Approximately 1 mg of the crude methanolic extract was dissolved in water and injected onto a ThermoElectron LTQ-Orbitrap high-resolution mass spectrometer with an Accela HPLC system using a C18 column (2.1 mm internal diameter ×10 cm length, 3 μm particle size, Ascentis). Mass spectra were recorded in positive mode using ESI. The following gradient was used at a flow rate of 200 ul/min: 10% methanol in water to 50% methanol over 30 min, followed by an increase to 100% methanol over 5 min. Both solvents contained 0.1% formic acid. Pure siderophores (approximately 10 μg) were used as standards using the same method. The L. pedicellatus extract was analyzed first, followed by washing of the column, a blank injection, and then the pure siderophore standard.
Bioinformatic analysis of NRPS domains
Module identification and domain organization of the NRPS was carried out using the online tools NRPS-PKS [30] and the PKS/NRPS analysis website [31]. Additional analyses utilized the software package HMMER [32] available from the website http://hmmer.org/. Alignments for Profile Hidden Markov Models (pHMMs) were downloaded from either the Pfam database [33] or the files supplied by Rausch et al. [34]. The hmmbuild command in HMMER was then used to build pHMMs.
Adenylation (A) domain: Protein sequences of A domains in TnbF were retrieved using the PKS/NRPS analysis website [31]. The specificity-conferring code of TnbF A domains were determined using the NRPSpredictor2 tool [35].
Condensation (C) domain: Protein sequences of C domains in TnbF were retrieved using the PKS/NRPS analysis website [31]. Aligned reference protein sequences of C domains were downloaded from Rausch et al. [34] and a subset of 91 sequences was used for the current study. The TnbF C domains were added to the downloaded C domain alignment using the multiple sequence alignment program MUSCLE [36]. The alignment was edited in Geneious v5.4 [37]. A maximum likelihood (ML) tree of C domains was reconstructed using RAxML 7.2.7 [38], implemented through the CIPRES portal [39]. The amino acid substitution matrix used in this analysis was the JTT matrix [40], with the Γ model of rate heterogeneity. RAxML's rapid bootstrap was performed with 100 replicates and the best scoring ML tree was saved.
Construction of tnbF plasmid insertion disruption mutant
For construction of the plasmids used for gene disruption, a region targeting the first C domain within tnbF was amplified with specific PCR primers TnbF851F and TnbF1193R (Table 1) and the high-fidelity Phusion DNA polymerase (Finnzymes). The addition of 3′ A-overhangs for TA cloning was carried out by DyNAzyme II DNA polymerase (Finnzymes) following the manufacturer's protocol. The amplicon was purified using the QIAquick PCR Purification Kit (Qiagen) and then cloned into pCR2.1 (Invitrogen). Plasmid DNA was isolated and double digested with XhoI and SacI. The resulting fragment was gel purified with the GENECLEAN II Kit (MP Biomedicals) and then ligated with the Quick Ligation Kit (NEB) into suicide vector pDM4, which had been double digested with the same restriction enzymes, resulting in plasmid pDMtnbF. The plasmid pDMtnbF was transformed into E. coli S17–1 λpir. This plasmid was then conjugated into T. turnerae T7901, and plasmid cointegrates were selected on SBM plates with 0.5% (w/v) Sigmacell cellulose Type 101 (Sigma) as the sole carbon source and 10 μg/ml chlorampenicol (Cm). The location of integration by pDMtnbF into the chromosome of T7901 was confirmed by PCR and DNA sequencing with vector-specific forward primers pNQ705 and pDM4CAT189 and the T7901 chromosomal-specific reverse primer TnbF1604R (Table 1). A list of all strains and plasmids used in this study are provided in Table 2.
Table 1. List of primers used in this study.
| Primer | Sequence (5′–3′) | Reference |
| TnbF851F | AAACCCTGGGAATGCCGTTTATGC | This study |
| TnbF1193R | TGCACGCCAAATTCAAAGTCGTCC | This study |
| TnbF1604R | TTTGCATAATGGCGAACATCGCGG | This study |
| pNQ705 | TTTGCGTAACGGCAAAAGCAC | Modified from Rock and Nelson, 2006 [64] |
| pDM4CAT189 | GAGCATTCATCAGGCGGGCA | This study |
Table 2. List of strains and plasmids used in this study.
| Strain or plasmid | Description | Reference |
| T. turnerae strains | ||
| T7901 | Wild-type | Distel et al., 2002 [21] |
| TtAH03 | tnbF::Cmr | This study |
| E. coli strains | ||
| TOP10 | F- mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu) 7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| S17–1 λpir | thi pro hsdR hsdM+ recA RP4–2-Tc::Mu-Km::Tn7λpir | Simon et al., 1983 [65] |
| Plasmids | ||
| pCR2.1 | T-vector, Kmr, Ampr | Invitrogen |
| pDM4 | Suicide vector, sacB gene, R6K origin, Cmr | Milton et al., 1996 [66] |
| pDMtnbF | Portion of tnbF gene cloned into pDM4 | This study |
Crude extracts from TtAH03 were analyzed and compared to those from wild-type strain T7901. Each strain was grown in iron-limited SBM medium with 0.5% (w/v) Sigmacell cellulose Type 101, with 10 μg/ml Cm in the case of TtAH03. Spent supernatant was extracted with HP20 resin. HP20 resin was washed with MilliQ water, 50% IPA, and 100% IPA. The 50% IPA fraction from each culture was dried by lyophilization and approximately 0.1 mg of each extract was used for HPLC analysis using the same method as described for compound isolation. Pure DHB was purchased from Sigma.
Results
Biosynthetic gene cluster
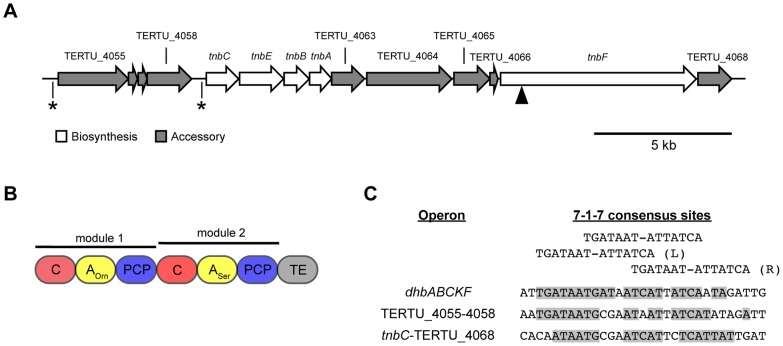
Annotation of the genome of Teredinibacter turnerae T7901 revealed a gene cluster with similarity to siderophore biosynthesis and iron transport genes [22] (Figure 1A). Proteins with closest similarity to the gene products of this cluster are shown in Table 3 and suggest the production of a catecholate siderophore. The genes tnbCEBA are homologous to the entCEBA genes, responsible for the biosynthesis and activation of 2,3-dihydroxybenzoate (DHB) via the shikimate pathway [41]. EntC isomerizes chorismate into isochorismate, then the N-terminal portion of EntB hydrolyzes isochorismate into 2,3-dihydro-DHB, which is oxidized to DHB by EntA. EntE then activates and transfers DHB to the aryl carrier C-terminal portion of EntB.
Figure 1. Overview of the turnerbactin biosynthetic gene cluster.
A. Organization of genes involved in turnerbactin biosynthesis. The locations of putative Fur boxes are indicated by asterisks (*). The black triangle indicates the location of NRPS disruption. B. Domain organization of the NRPS modules of TnbF. C, condensation domain; A, adenylation domain; PCP, peptidyl-carrier protein; TE, thioesterase. C. Putative Fur box sequences of turnerbactin's biosynthetic gene cluster compared to the bacillibactin biosynthetic operon of Bacillus subtilis. Shaded bases match the revised view of Fur box sequences consisting of two overlapping 7–1–7 motifs proposed by Baichoo and Helmann.
Table 3. Proteins with similarity to the products of the turnerbactin biosynthetic gene cluster. S: similarity, I: identity.
| Gene | Protein size | Proposed function | Homolog (Accession, organism) | S/I (%) |
| TERTU_4055 | 648 | TonB-dependent receptor | CCD03052, Azospirillum brasilense Sp245 | 68/48 |
| TERTU_4056 | 87 | Hypothetical protein | ABL99859, Shewanella amazonensis | 51/37 |
| TERTU_4057 | 100 | Hypothetical protein | EGM6880, Shewanella sp. HN-41 | 72/56 |
| TERTU_4058 | 550 | PepSY-associated TM helix domain protein | ABK46690, Shewanella sp. ANA-3 | 64/48 |
| tnbC | 394 | Isochorismate synthase | AAN33228 Brucella suis 1330 | 66/51 |
| tnbE | 538 | 2,3-dihydroxybenzoate-AMP ligase | EHK70948 Pseudomonas psychrotolerans L19 | 76/61 |
| tnbB | 292 | Isochorismatase | CAG23680 Photobacterium profundum SS9 | 73/54 |
| tnbA | 255 | 2,3-dihydroxybenzoate-2,3-dehydrogenase | EHK70946 Pseudomonas psychrotolerans L19 | 72/58 |
| TERTU_4063 | 389 | RND family efflux transporter | ABE56876 Shewanella denitrificans OS217 | 66/44 |
| TERTU_4064 | 1047 | HAE1 family RND transporter | ABC30520 Hahella chejuensis KCTC 2396 | 76/60 |
| TERTU_4065 | 445 | Esterase | ACS85292 Dickeya dadantii Ech703 | 50/38 |
| TERTU_4066 | 85 | MbtH domain protein | ACZ77654 Dickeya dadantii Ech586 | 73/56 |
| tnbF | 2399 | NRPS | AAN28936 Acinetobacter baumannii | 62/45 |
| TERTU_4068 | 417 | Enterobactin exporter | ZP_10160264 Vibrio campbellii DS40M4 | 73/52 |
The gene tnbF codes for a 2399 amino acid, two module NRPS. Each module contains a C, A, and peptidyl-carrier protein (PCP) domain, followed by a C-terminus thioesterase (TE) domain (Figure 1B). A phosphopantetheinyl (P-pant) transferase, which is required for NRPS biosynthesis [42], was not found at this locus. The genome of T. turnerae T7901 codes for two P-pant transferases, TERTU_1510 and TERTU_4652. Due to its homology to the entD gene of enterobactin biosynthesis, the gene product of TERTU_1510 is proposed to be the P-pant transferase that posttranslationally attaches a P-pant moiety to a conserved Ser residue of the apo-PCP domains of TnbF.
In addition to biosynthesis genes, a number of other genes are also found in this cluster that are likely to assist in siderophore transport and uptake. TERTU_4066 codes for an MbtH-like protein [43], a protein of unknown function that is found in many but not all NRPS gene clusters. TERTU_4068 codes for a putative homologue of EntS, a 12 trans-membrane domain-containing efflux pump belonging to the Major Facilitator Superfamily (MFS) shown to excrete enterobactin in Escherichia coli [44]. TERTU_4055 codes for a putative TonB-dependent receptor, required for the recognition and uptake of the ferric siderophore complex. TERTU_4065 shares similarity to the gene encoding the enterobactin esterase, fes. In E. coli, once ferric enterobactin has entered the cell, iron is removed from the siderophore by hydrolyzing the ester bonds of the siderophore's backbone with the fes gene product [45].
This gene cluster also shows two putative ferric uptake regulator (Fur) box sequences (Figure 1C), suggesting transcriptional regulation by one of the three Fur homologs found in the genome, TERTU_0053, TERTU_3299, and TERTU_3389. These sequences are compared to the bacillibactin biosynthetic operon of Bacillus subtilis in Figure 1C, whose Fur box matches exactly to the classic Fur box sequence proposed by de Lorenzo et al. [46]. The Fur box sequences of turnerbactin's biosynthetic gene cluster show high similarity with the two overlapping 7–1–7 motifs proposed by Baichoo and Helmann [47]. This similarity suggests that the turnerbactin biosynthetic gene cluster is composed of two iron-regulated operons: the first operon consisting of the genes TERTU_4055–4058, and the second operon consisting of the genes tnbC-TERTU_4068. Support for the role of iron in regulating expression of this gene cluster was provided by assaying CAS activity of iron-limited and iron-replete cultures (Figure 2). The high CAS activity observed in iron-limited cultures in conjunction with the negligible activity observed in iron-replete conditions suggests that low iron conditions leads to increased production of the siderophore.
Figure 2. Fe(III)-binding activity of T. turnerae T7901 cultures.
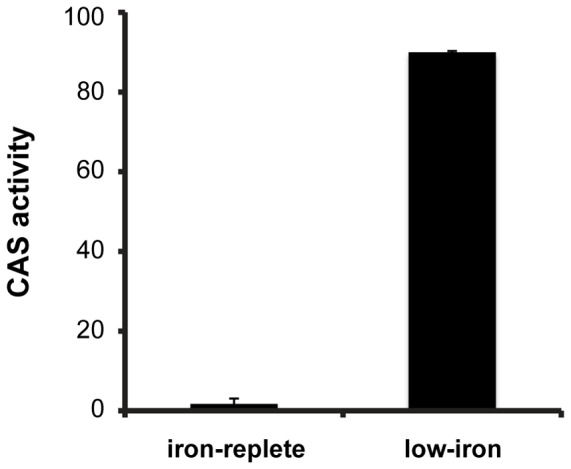
The CAS assay was used to measure Fe(III)-binding activity of iron-replete and iron-limited culture supernatants. Iron-replete conditions contained 10 μM ferric iron, while iron-limited conditions contained 0.1 μM ferric iron.
NRPS domain specificities
The crystal structure of the Phe-activating A domain of gramicidin synthetase A (GrsA) by Conti et al. [48] identified 10 residue positions critical in substrate binding. Stachelhaus et al. [49] and Challis et al. [50] determined that these 10 residues correspond to similar residues in other A domains and that these residues would form a “specificity-conferring code” that could be used to infer specificity of uncharacterized A domains. The amino acid specificity of A domains involved in turnerbactin biosynthesis was analyzed by comparing their respective active-site residues to those of known A domains.
The online tool NRPSpredictor2 [35] was used to determine specificity-conferring codes. The specificity-conferring code of TnbE, believed to be involved in 2,3-DHB biosynthesis and activation, is PLPAQGVVNK, which matches exactly that of DhbE, the homologue in bacillibactin biosynthesis. The specificity-conferring code of the second module (M2) A domain was determined to be DVWHFSLVDK, matching exactly with that of the Ser-activating A domain of the enterobactin synthetase as well as several other characterized Ser-activating A domains. However, the prediction for the first module (M1) A domain was inconclusive, as the specificity-conferring code of DSDDGGLVDK did not match any previously characterized specificity-conferring codes. The closest matches to known specificity-conferring codes at 60% were that of the M1 A domain of the chrysobactin synthetase (CbsF) and the M1 A domain of the vanchrobactin synthetase (VabF) which activate Lys and Arg, respectively. BLAST analysis of the entire M1 A domain showed highest similarity to the M1 A domains of CbsF and VabF at 65%/44% (similarity/identity) and 59%/42%, respectively.
In addition to A domain specificity prediction, C domains can also be subject to bioinformatic prediction. Rausch et al. [34] demonstrated that the reconstructed phylogeny of C domains shows a grouping according to function rather than species phylogeny or substrate specificity. The results of a maximum likelihood phylogenetic analysis with TnbF's C domains show that the TnbF M1 C domain groups with other Starter domains, as expected from its location in the NRPS (Figure S1). The M2 C domain of TnbF groups within the DCL functional domains and this placement is strongly supported, evidenced by the high bootstrap values at the base of the Dual E/C and DCL groups. In most cases, DCL domains are preceded by an epimerase (E) domain, catalyzing the epimerization of amino acids from L to D configuration. However, an E domain was not detected in TnbF through pHMM analysis and an external racemase was not detected in the gene cluster. In conjunction with the chemical analysis of the siderophore product (below), this suggests that the M2 C domain of TnbF exhibits LCL activity as opposed to its predicted DCL activity.
Chemical characterization of the siderophore
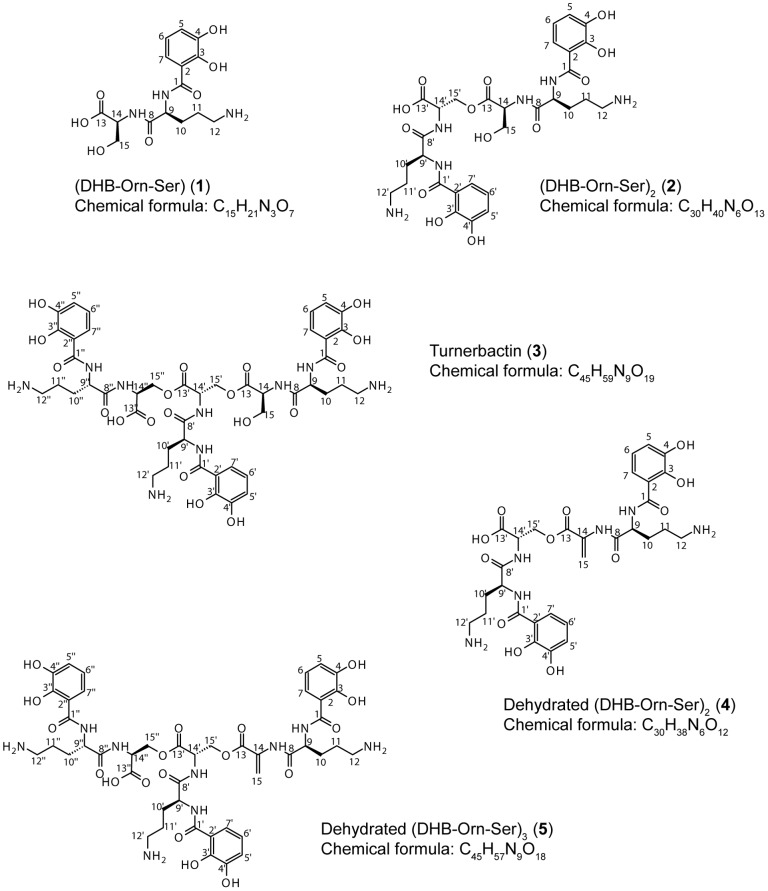
The structures of the siderophores isolated from T. turnerae T7901 are shown in Figure 3. The siderophores from T. turnerae were purified from iron-deficient liquid SBM cultures by adsorbing siderophores from culture supernatants on an HP20 column, followed by purification by RP-HPLC. The CAS assay was used to track the siderophores throughout the purification process. RP-HPLC revealed five peaks displaying CAS activity (Figure S2). High resolution electrospray ionization mass spectrometry (HRESIMS) determined the mass of the molecular ion [M+H]+: DHB-Orn-Ser (1), m/z 356.1454, corresponding to a molecular formula of C15H22N3O7 (calculated 356.1452); (DHB-Orn-Ser)2 (2), m/z 693.2736, corresponding to a molecular formula of C30H41N6O13 (calculated 693.2726); turnerbactin (3), m/z 1030.4003, corresponding to a molecular formula of C45H60N9O19 (calculated 1030.4000); dehydro-(DHB-Orn-Ser)2 (4), m/z 675.2626, corresponding to a molecular formula of C30H39N6O12 (calculated 675.2620); dehydro-(DHB-Orn-Ser)3 (5), m/z 1012.3905, corresponding to a molecular formula of C45H58N9O18 (calculated 1012.3894).
Figure 3. Structures of siderophores isolated from T. turnerae T7901.
ESIMS/MS analysis of each of these compounds is summarized in Table 4. All compounds exhibited similar fragmentation patterns, with overlap of fragments when applicable. The masses of the fragments could be correlated with the loss of various constituents of the siderophore (Table 4).
Table 4. Molecular ions and common mass fragments of siderophore from T. turnerae T7901. Fragment losses refer to the compound listed immediately above in the table.
| Dehydrated (DHB-Orn-Ser)3 (5) | Dehydrated (DHB-Orn-Ser)2 (4) | Turnerbactin (3) | (DHB-Orn-Ser)2 (2) | (DHB-Orn-Ser) (1) | Fragment |
| 1012.4 | 675.2 | 1030.4 | 693.3 | 356.1 | Parent ion |
| 762.3 | 780.3 | Loss of DHB-Orn | |||
| 675.2 | 693.3 | 675.2 | Loss of Ser | ||
| 425.2 | 425.2 | 443.2 | 443.2 | Loss of DHB-Orn | |
| 338.1 | 338.1 | 356.1 | 356.1 | Loss of Ser | |
| 251.1 | 251.1 | 251.1 | 251.1 | 251.1 | DHB-Orn |
| 115.1 | 115.1 | 115.1 | 115.1 | 115.1 | Orn |
The 1–D 1H and 13C NMR assignments of 1 were confirmed by 2–D 1H-1H TOCSY, HSQC, and HMBC experiments (Table 5, Figures S4-S14). The 13C NMR spectrum shows 15 distinct C resonances corresponding to three carbonyl carbons (δ 169.98 to 172.12), four methylene carbons (δ 23.39 to 38.93 for ornithine, δ 61.35 for serine), two methine carbons (δ 52.26 for ornithine, δ54.82 for serine), and six aromatic carbons (δ 116.01 to 147.81). The 1H NMR spectrum shows 12 distinct resonances corresponding to three aromatic protons (δ 6.77 to 7.36), seven methylene protons (δ 1.79 to 3.00 ornithine, δ 3.88, 3.98 for serine), and two methine protons (δ 4.82 for ornithine, δ 4.55 for serine). The aromatic splitting pattern in the 1H NMR spectrum is indicative of a 2,3-DHB moiety. A HMBC correlation between the α-proton of ornithine and the carbonyl carbon of DHB indicates that DHB is attached to the α-amine group of ornithine. A HMBC correlation between the α-proton of serine and the carbonyl carbon of ornithine confirms the ornithine-serine peptide bond.
Table 5. NMR data for 1 and 3 (800 MHz) in CD3OD.
| (DHB-Orn-Ser) (1) | Turnerbactin (3) | ||||||
| Position | δC | δH (J in Hz) | TOCSY | HMBC | δH (J in Hz) | HSQC | HMBC |
| DHB | |||||||
| 1, 1′, 1′′ | 169.98 | 169.1 | |||||
| 2, 2′, 2′′ | 116.01 | 115.9 | |||||
| 3, 3′, 3′′ | 147.81 | 147.7 | |||||
| 4, 4′, 4′′ | 145.82 | 145.7 | |||||
| 5, 5′, 5′′ | 118.38 | 6.98, dd (1.6, 8.0), [1H] | 6, 7 | 2, 3, 4, 6, 7 | 6.97, m, [3H] | 118.5 | 2, 3, 4, 6, 7 |
| 6, 6′, 6′′ | 118.52 | 6.77, t (8.0), [1H] | 5, 7 | 1, 2, 3, 4, 5, 7 | 6.76, m, [3H] | 118.7 | 1, 2, 3, 4, 5, 7 |
| 7, 7′, 7′′ | 118.44 | 7.36, dd (0.8, 8.0), [1H] | 5, 6 | 1, 2, 3, 4, 5, 6 | 7.36, m, [3H] | 118.6 | 1, 2, 3, 4, 5, 6 |
| Ornithine | |||||||
| 8, 8′, 8′′ | 172.12 | 172.2 | |||||
| 9 | 52.26 | 4.82, dd (5.6, 8.0), [1H] | 10, 11, 12 | 1, 8, 10, 11 | 4.71, m, [1H] | 52.6 | 1, 8, 10, 11 |
| 9′ | 4.77, m, [1H] | 52.5 | 1′, 8′, 10′, 11′ | ||||
| 9′′ | 4.79, m, [1H] | 52.2 | 1′′, 8′′, 10′′, 11′′ | ||||
| 10, 10′, 10′′ | 29.06 | 2.09, m [1H]; 1.89, m [1H] | 9, 10, 11, 12; 9, 10, 11, 12 | 8, 9, 11, 12 | 2.05, m, [3H]; 1.89, m, [3H] | 28.7 | 8, 9, 11, 12; 8, 9, 11, 12 |
| 11, 11′, 11′′ | 23.39 | 1.83, m, [1H]; 1.79, m [1H] | 9, 10, 11, 12; 9, 10, 11, 12 | 9, 10, 12 | 1.81, m, [6H] | 28.14, 28.13 | 9, 10, 12 |
| 12, 12′, 12′′ | 38.93 | 3.00, m, [2H] | 9, 10, 11, 12 | 10, 11 | 2.99, m, [6H] | 38.9 | 10, 11 |
| Serine | |||||||
| 13 | 171.91 | 169.8 | |||||
| 13′ | 168.6 | ||||||
| 13′′ | 168.8 | ||||||
| 14 | 54.82 | 4.55, t (8.0,) [1H] | 15 | 13, 15 | 4.56, m, [1H] | 55.0 | 8, 13, 15 |
| 14′ | 4.80, m, [1H] | 52.0 | 8′, 13′, 15′ | ||||
| 14′′ | 4.87, m, [1H] | 51.7 | 8′′, 13′′, 15′′ | ||||
| 15 | 61.35 | 3.98, dd (4.8, 11.2), [1H]; 3.88, dd (4.0, 11.2), [1H] | 14, 15 | 13, 14 | 3.95, m, [1H]; 3.82, m, [1H] | 62.2 | 13, 14; 13, 14 |
| 15′ | 4.72, m, [1H]; 4.43, m, [1H] | 64.5 | 13, 13′, 14′; 13, 13′, 14′ | ||||
| 15′′ | 4.57, m, [1H]; 4.49, m, [1H] | 63.5 | 13′′, 14′′; 13′′, 14′′ | ||||
The structures of 2 and 3 were inferred using MS, 1H NMR, and 2–D NMR experiments HSQC and HMBC (2: Table S1, Figures S15-S20; 3: Table 5, Figures S21-S26). The 1H NMR spectra of 2 and 3 are similar to that of 1, with the addition of a downfield shift of serine methylene protons of 2 and 3 (δ 4.43 to 4.75) compared to the serine methylene protons of 1 (δ 3.88, 3.98), indicating the presence of serine ester linkages in 2 and 3.
The MS parent ion masses of 5 suggested a cyclic trimer as the result of an additional serine ester bond, leading to a triserine lactone backbone. However, 1H and 13C NMR in addition to 2-D experiments 1H-1H TOCSY, HSQC, and HMBC revealed a dehydrated analogue of the linear trimer (Table S1, Figures S37-S46). The dehydration of the serine hydroxyl group resulted in an alkene, corresponding to a carbon resonance of δ 112.81 and proton resonances of δ 6.12 and 5.80. Similar spectra were obtained for 4 (Table S1, Figures S27-S36).
Chiral amino acid analysis of turnerbactin was carried out using Marfey's method [29]. The hydrolysate of turnerbactin was derivatized with FDAA and compared to amino acid standards derivatized in the same manner (Figure S3), revealing the presence of L-ornithine and L-serine.
A UV-visible spectrum of purified turnerbactin showed an absorption peak at 330 nm, characteristic of catecholate siderophores.
Disruption of tnbF diminishes Fe binding activity
In order to correlate the tnb gene cluster with the biosynthesis of turnerbactin, a tnbF mutant was constructed by integrating a chloramphenicol (Cm) resistance cassette into the M1 C domain of tnbF by a single crossover recombination (Figure 1A). The resulting strain was named TtAH03.
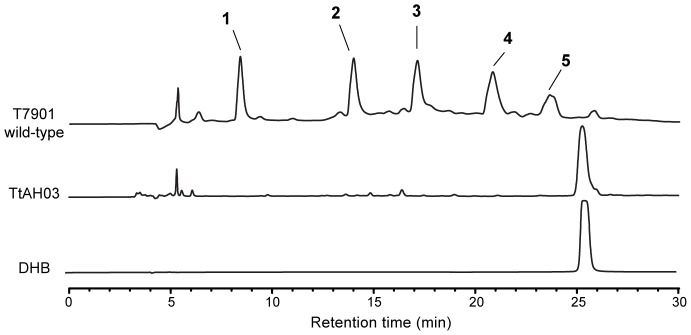
Disruption of tnbF resulted in a significant reduction in Fe(III)-binding activity as measured by the CAS assay compared to wild-type strain T7901 (Figure 4). The residual CAS activity shown for TtAH03 is presumably due to the DHB units produced by the upstream genes tnbCEBA. For further support, crude culture extracts from both wild-type T7901 and mutant TtAH03 were compared by HPLC, alongside pure DHB (Figure 5). The extract of wild-type T7901 shows the presence of all five siderophores, as expected. TtAH03 lacks these five peaks while showing a prominent peak at about 25.5 min, corresponding to the retention time of pure DHB. These results support the proposed role of the tnb gene cluster in the role of turnerbactin biosynthesis.
Figure 4. Fe(III)-binding activity of wild-type T. turnerae T7901 compared to tnbF mutant TtAH03.
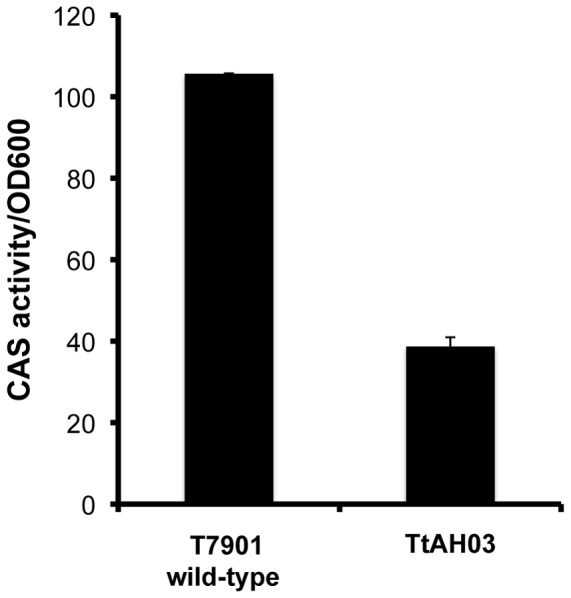
Disruption of tnbF leads to a significant decrease in siderophore activity, as measured by the CAS assay. CAS activity was normalized to OD600 measurements of each culture.
Figure 5. Comparison of wild-type T7901 and TtAH03 extracts by HPLC, recorded at 215 nm.
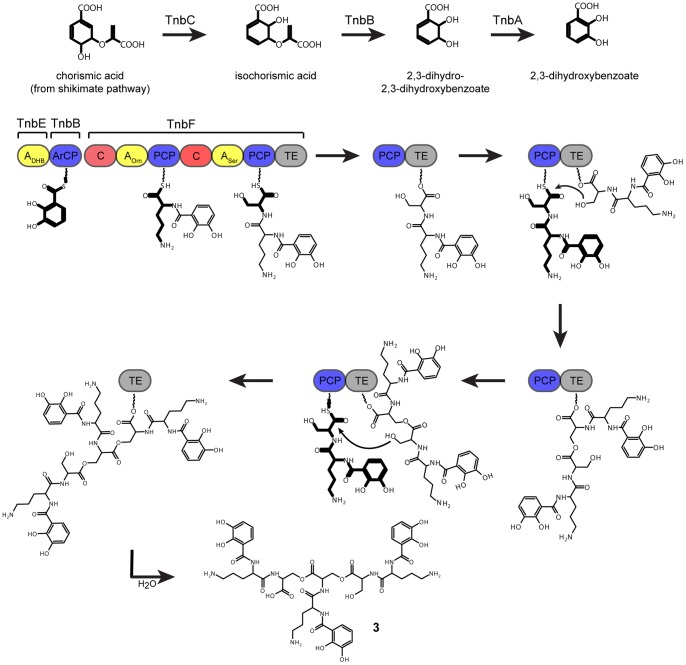
Biosynthesis of turnerbactin
The proposed biosynthesis is shown in Figure 6. DHB is activated by TnbE and then transferred to the aryl carrier C-terminal portion of TnbB. L-Orn is activated by the M1 A domain of TnbF and the M1 C domain condenses DHB to Orn, forming a DHB-Orn intermediate on the M1 PCP domain. L-Ser is activated by the M2 A domain and the M2 C domain condenses the DHB-Orn intermediate to the Ser to form the thioester intermediate DHB-Orn-Ser-S-PCP on the M2 PCP domain. A conserved Ser residue in the TE domain mounts a nucleophilic attack on the PCP domain-bound thioester intermediate, leading to a DHB-Orn-Ser-O-TE ester intermediate. After another round of DHB-Orn-Ser biosynthesis, the Ser side chain hydroxyl group of the DHB-Orn-Ser-O-TE intermediate attacks the DHB-Orn-Ser-S-PCP to yield a (DHB-Orn-Ser)2-S-PCP, which is then transferred to the TE domain. A third consecutive round yields a (DHB-Orn-Ser)3-O-TE intermediate, which is then hydrolyzed by water to yield the linear trimer (DHB-Orn-Ser)3, turnerbactin (3). It is uncertain whether the other compounds (1, 2, 4, and 5) discovered in this study are byproducts from regulated biosynthesis by the NRPS, incomplete biosynthesis by the NRPS, enzymatic degradation by an esterase, or hydrolysis products from either the purification process or culture conditions.
Figure 6. Proposed biosynthesis of turnerbactin.
Bold structures indicate the most recent addition to the compound.
Detection of turnerbactin in shipworm samples
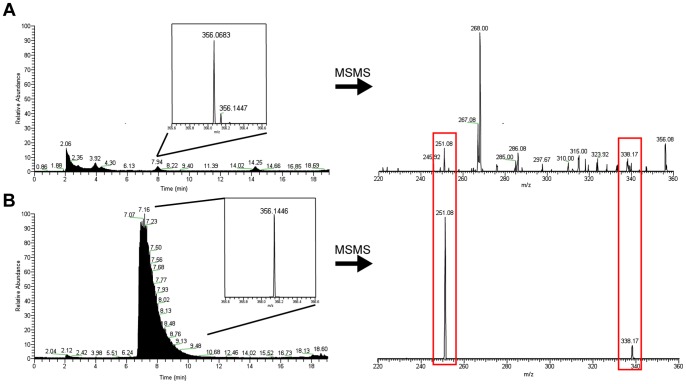
The potential for the involvement of turnerbactin in the symbiosis between T. turnerae and its shipworm host was examined. Detection of the compound in the shipworm host would provide evidence that the compound is produced and utilized in the symbiosis. The presence of turnerbactin and the related DHB-Orn-Ser products in the shipworm host was investigated using MS. The shipworm, Lyrodus pedicellatus, is known to contain the symbiont T. turnerae [51], [52]. A crude, methanolic extract of whole L. pedicellatus tissue samples was analyzed by LC-HRESIMS/MS and compared to pure siderophores isolated from T. turnerae T7901 as standards. The method used in this analysis shows pure DHB-Orn-Ser eluting from the column over the span of approximately 7.5–8.5 min (Figure 7). The HRESIMS m/z peak for this elution product is 356.1446. MS/MS analysis shows the presence of two daughter ions with m/z values of 338.17 and 251.08. Analysis of the L. pedicellatus extract shows a peak eluting at 7.94 min with an m/z value of 356.1447. MS/MS analysis of this peak shows the presence of the two daughter ions at 338.17 and 251.08. In order to discount contamination of the column, L. pedicellatus extracts were run before siderophore standards with a blank injection before each run. Turnerbactin and the other siderophore fragments were not detected in L. pedicellatus extracts. The lack of detection of these products may be due to degradation of these products or they may have been present in concentrations below the detection limit of the assay.
Figure 7. HPLC, HRMS, and MS/MS analysis of L. pedicellatus extracts.
A. Extract of L. pedicellatus. B. Standard of DHB-Orn-Ser, isolated from T. teredinibacter T7901. Inset in both figures shows the HRMS of the peak indicated. The shipworm extract shows other compounds with similar mass, but also contains a compound with nearly identical high resolution mass as the DHB-Orn-Ser standard of 356.1446. Red boxes indicate shared MS/MS fragmentation peaks of the 356 molecular ion between the shipworm extract and the siderophore standard.
Discussion
This work describes the purification, structural characterization, and biosynthesis genes of a novel siderophore, turnerbactin, from the shipworm symbiont T. turnerae T7901. Turnerbactin was isolated along with a monomer, dimer, dehydrated dimer, and dehydrated trimer of DHB-Orn-Ser. We propose that turnerbactin is produced beginning with the production and activation of DHB by the protein products of tnbCEBA. The NRPS encoded by tnbF then activates and condenses first L-Orn to DHB and then L-Ser to DHB-L-Orn. Turnerbactin is likely assembled by tnbF in such a way that the modules are used three consecutive times to produce a trimer of 2,3-DHB-L-Orn-L-Ser before being hydrolytically released by the C-terminal TE domain to yield the linear peptide product.
The cyclic form of turnerbactin was not found in this study. Because previously reported trimeric triscatecholate siderophores have been found in their cyclic form (enterobactin [9], [10], bacillibactin [12], cyclic trichrysobactin [15], streptobactin [16]), with the exception of the recently reported linear trivanchrobactin [18], we wanted to ensure that the lack of the cyclic form of turnerbactin was not an artifact of the purification method. Similarly, we wanted to ensure that the dehydroalanine (Dha) residue resulting from a dehydrated Ser residue, which has not been previously reported in siderophores, was also not an artifact of the purification method. To determine if the lack of a cyclic trimer and the finding of dehydrated products were artifacts of the purification method a control experiment with cyclic trichrysobactin from D. chrysanthemi [15] was carried out. The monomeric forms of chrysobactin and turnerbactin both contain a Ser backbone with a DHB functional group and differ only in the spacer amino acid: D-lysine in chrysobactin, L-ornithine in turnerbactin. A side-by-side purification with the same method yielded cyclic trichrysobactin, trichrysobactin, dichrysobactin, chrysobactin, and no dehydrated products from D. chrysanthemi while yielding the same suite of siderophores as described in this study from T. turnerae. Due to their high degree of structural similarity, this control experiment suggests that the lack of a cyclic trimer is indeed not an artifact of the purification method and the dehydration may be due to an enzymatic modification on the part of T. turnerae.
A Dha residue is not unprecedented in other biosynthesized compounds. The formation of Dha residues has been previously reported in ribosomally-produced (RP) compounds such as lantibiotics [53] and thiopeptide antibiotics [54], and in non-RP compounds such as the microcystins [55], [56], [57]. The LanB family of enzymes catalyzes the dehydration of amino acids in the biosynthesis of RP compounds. Using BLAST and pHMM analysis, a potential homolog of this enzyme was not found in the genome of T. turnerae T7901. The exact mechanism of non-RP formation of Dha is unclear. However, an enzyme in the biosynthetic pathway of microcystin in Microcystis aeruginosa, McyI, with similarity to D-3-phosphoglycerate dehydrogenases, is thought to be responsible for the dehydration of Ser in microcystin [57]. BLAST analysis of McyI against the genome of T. turnerae T7901 did identify a D-3-phosphoglycerate dehydrogenase, encoded by the gene TERTU_0393 (S/I, 50%/32%), distantly located from the turnerbactin biosynthetic gene cluster. However, since this enzyme is essential for serine biosynthesis, it is difficult to implicate this enzyme in the dehydration of Ser in turnerbactin. More broadly, a dehydrated amino acid in a siderophore has been previously reported in the loihichelins [58]. The loihichelins are a suite of amphiphilic siderophores from the marine bacterium Halomonas sp. LOB-5 and contain dehydroamino-2-butyric acid. However, a mechanism for this dehydration has not been reported. Alternatively, the dehydration of Ser in turnerbactin may occur through a non-enzymatic means, though as noted above, the control experiment with cyclic trichrysobactin suggests that this does not occur during the purification process.
The bioinformatic analyses of tnbF's catalytic domains suggest these domains elude accurate prediction. Predictive analysis of the M1 A domain failed to suggest a specificity with reasonable cut-off scores. C domain analysis suggests a DCL acting domain, while chemical data suggest a LCL acting domain. Taken together, the addition of tnbF to the databases of NRPS domains may help to improve the functional prediction of as-yet-undiscovered NRPS domains.
The NRPS's responsible for the production of glycopeptide antibiotics, found in actinomycetes, also exhibit aberrant C domain prediction. Glycopeptide NRPS's contain seven modules. Rausch et al. [34] showed that the M4 and M7 C domains act as LCL domains while clustering in the DCL group of C domains in phylogenetic analysis, similar to the M2 C domain of TnbF (Figure S1). While the M2 C domain of TnbF does not cluster directly with these glycopeptide C domains, a similar change of function is assumed to have occurred, most likely a result of convergent evolution. Occasionally, an external racemase can be found in a biosynthetic gene cluster, providing a D-amino acid for the NRPS. This is the case for cyclosporine, where an external racemase provides D-Ala for the first module of the cyclosporine synthetase [59]. However, an external racemase was not detected in the turnerbactin biosynthetic gene cluster and D-Orn was not detected in amino acid analysis.
Nevertheless, turnerbactin represents a novel siderophore, structurally similar to the catecholate siderophores trivanchrobactin and trichrysobactin. A catecholate siderophore was partially characterized from the soil diazotroph, Azospirillum brasilense, which contained equimolar amounts of 2,3-DHB, Orn, and Ser [60]. However, a structure was neither elucidated nor presented. Turnerbactin, vanchrobactin, and chrysobactin share a 2,3-DHB functional moiety, a Ser backbone, and a hydrophilic long, positively charged spacer amino acid. The latter trait distinguishes these siderophores from the widely studied triscatecholate enterobactin. However, the biosynthesis of turnerbactin differs from vanchrobactin and chrysobactin in that the spacer amino acid is not epimerized to a D-configuration, due to the absence of an E domain in TnbF. The similarity of TnbF to the vanchrobactin and chrysobactin NRPS's and the finding of a DCL-like C domain for the M2 of TnbF suggest that an E domain may have been lost at some point.
The recent reports of trivanchrobactin [18], cyclic trichrysobactin [15], and streptobactin [16], along with the current reporting of turnerbactin, add to the growing family of triscatecholate siderophores. As mentioned previously, the triscatecholate siderophores contain the highest stability constants for Fe(III) measured for siderophores to date [9], [10]. When comparing a cyclic compound to its linear counterpart, a cyclic compound will likely have a higher stability constant, as the flexibility of the cyclic ligand and its corresponding iron complex will be less than that of the linear ligand, thereby decreasing the entropy difference. This is seen in the stability constants for cyclic enterobactin (1049) compared to its linearized form, the linear trimer of DHB-serine (1043) [61]. However, the linear trimer still provides a siderophore with relatively strong affinity for iron and the initial rates of uptake for both the linear trimer and linear dimer ferric enterobactin complexes are essentially the same as the cyclic ferric enterobactin complex [61]. The role of iron in regulating the production of turnerbactin is supported by the identification of two putative Fur box sequences in the biosynthetic gene cluster and the increased Fe(III)-binding activity of T. turnerae T7901 cultures in response to iron-limited conditions.
A fragment of turnerbactin, DHB-Orn-Ser (1), was detected in extracts of the shipworm L. pedicellatus. The compound detected in the shipworm shared the same LC retention time, high-resolution molecular ion mass, and daughter ions in tandem MS fragmentation as a pure DHB-Orn-Ser standard. The detection of this monomer unit in a shipworm sample suggests that turnerbactin is produced and utilized in some capacity in the symbiosis. Since whole animal tissue was used in these experiments, it is not known whether these siderophores are confined to the immediate vicinity of the symbionts in the gills, or if they appear in other locations of the shipworm. The exact role of endosymbiont-derived siderophores in the host is currently unclear. The caecum of L. pedicellatus is largely devoid of microbes [62]. The mechanism by which this occurs is currently unknown. It has been proposed that secondary metabolites produced by gill endosymbionts such as T. turnerae may be translocated to the caecum and contribute to suppression of microbial growth [24]. Siderophores may also play a role in suppression of microbial growth in the caecum. Studies in plant systems have found that siderophores can play a role in nutrient deprivation [63]. By sequestering the iron, siderophores can make iron unavailable to competing microbes, thereby restricting their growth. This theory could be tested by MS-based screening of dissected organs and tissues of shipworms samples. In addition, chemical localization studies using methods such as MS-imaging would allow the localization of the siderophore within host tissue. These types of studies would be valuable in determining the extent to which siderophores are utilized in the host and lend insight into their possible roles, such as intersymbiont competition or suppressing competing microbes in the caecum.
Supporting Information
Maximum likelihood tree of C domains, showing the grouping of different C domain subtypes. The C domains of TnbF are shown in red. TnbF's M2 C domain groups with the DCL functional group, while the M1 C domain groups with the Starter functional group. Each C domain is labeled with the organism name, followed by accession number, followed by the module number from which the C domain is referring. C domains from glycopeptide antibiotic NRPS's are shown in blue. Bootstrap values are based on 100 replicates and are only shown for the basal branches of groups.
(TIF)
HPLC trace of HP20 extract from T. turnerae T7901 culture supernatant, recorded at 215 nm.
(TIF)
HPLC trace of derivatized hydrolysate of turnerbactin and chiral amino acid standards, recorded at 340 nm. (A) hydrolysate of turnerbactin, (B) L-ornithine, (C) DL-ornithine, (D) L-serine, (1) D-ornithine, (2) L-ornithine, (3) L-serine, (4) Marfey's reagent (FDAA).
(TIF)
1 1H NMR spectrum (800 MHz) in CD3OD.
(TIF)
1 13C NMR spectrum (800 MHz) in CD3OD.
(TIF)
1 1H-13C HSQC spectrum (800 MHz) in CD3OD.
(TIF)
1 1H-13C HMBC spectrum (800 MHz) in CD3OD.
(TIF)
1 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
1 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
1 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
1 1H-1H TOCSY spectrum (800 MHz) in CD3OD.
(TIF)
1 1H-1H TOCSY spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
1 1H-1H TOCSY spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
1 1H-1H TOCSY spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
2 1H NMR spectrum (800 MHz) in CD3OD.
(TIF)
2 1H-13C HSQC spectrum (800 MHz) in CD3OD.
(TIF)
2 1H-13C HMBC spectrum (800 MHz) in CD3OD.
(TIF)
2 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
2 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
2 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
3 1H NMR spectrum (800 MHz) in CD3OD.
(TIF)
3 1H-13C HSQC spectrum (800 MHz) in CD3OD.
(TIF)
3 1H-13C HMBC spectrum (800 MHz) in CD3OD.
(TIF)
3 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
3 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
3 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
4 1H NMR spectrum (800 MHz) in CD3OD.
(TIF)
4 13C NMR spectrum (800 MHz) in CD3OD.
(TIF)
4 1H-13C HMBC spectrum (800 MHz) in CD3OD.
(TIF)
4 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
4 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
4 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
4 1H-1H TOCSY spectrum (800 MHz) in CD3OD.
(TIF)
4 1H-1H TOCSY spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
4 1H-1H TOCSY spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
4 1H-1H TOCSY spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
5 1H NMR spectrum (800 MHz) in CD3OD.
(TIF)
5 13C NMR spectrum (800 MHz) in CD3OD.
(TIF)
5 1H-13C HMBC spectrum (800 MHz) in CD3OD.
(TIF)
5 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
5 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
5 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
5 1H-1H TOCSY spectrum (800 MHz) in CD3OD.
(TIF)
5 1H-1H TOCSY spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
5 1H-1H TOCSY spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
5 1H-1H TOCSY spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
NMR data for (DHB-Orn-Ser)2 (2), Dehydrated (DHB-Orn-Ser)2 (4), and Dehydrated (DHB-Orn-Ser)3 (5) (800 MHz) in CD3OD.
(PDF)
Acknowledgments
We thank Cheryl Hodson Shirley, Dr. David Peyton, and Dr. Hongjun Zhou for technical assistance with NMR data acquisition, and Dr. Zhenjian Lin, Dr. Eric Schmidt, and Dr. Hiroaki Naka for helpful discussions.
Funding Statement
1. Agency: United States National Institutes of Health, Grant number: 1U01TW008163 (URL: http://grants.nih.gov/grants/oer.htm); 2. Agency: United States National Science Foundation, Grant number: 1059067 (URL: http://www.nsf.gov/funding/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rue EL, Bruland KW (1995) Complexation of iron(III) by natural organic ligands in the Central North Pacific as determined by a new competitive ligand equilibration/adsorptive cathodic stripping voltammetric method. Marine Chemistry 50: 117–138. [Google Scholar]
- 2. Wu J, Luther GW (1995) Complexation of Fe(III) by natural organic ligands in the Northwest Atlantic Ocean by a competitive ligand equilibration method and a kinetic approach. Marine Chemistry 50: 159–177. [Google Scholar]
- 3. Ratledge C, Dover LG (2000) Iron metabolism in pathogenic bacteria. Annu Rev Microbiol 54: 881–941. [DOI] [PubMed] [Google Scholar]
- 4. Chu BC, Garcia-Herrero A, Johanson TH, Krewulak KD, Lau CK, et al. (2010) Siderophore uptake in bacteria and the battle for iron with the host; a bird's eye view. Biometals 23: 601–611. [DOI] [PubMed] [Google Scholar]
- 5. Raymond KN, Muller G, Matzanke BF (1984) Complexation of iron by siderophores a review of their solution and structural chemistry and biological function. Topics in Current Chemistry 123: 49–102. [Google Scholar]
- 6. Winkelmann G (2002) Microbial siderophore-mediated transport. Biochem Soc Trans 30: 691–696. [DOI] [PubMed] [Google Scholar]
- 7. Sandy M, Butler A (2009) Microbial iron acquisition: marine and terrestrial siderophores. Chem Rev 109: 4580–4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Challis GL (2005) A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. ChemBioChem 6: 601–611. [DOI] [PubMed] [Google Scholar]
- 9. O'Brien IG, Gibson F (1970) The structure of enterochelin and related 2,3-dihydroxy-N-benzoylserine conjugates from Escherichia coli . Biochim Biophys Acta 215: 393–402. [DOI] [PubMed] [Google Scholar]
- 10. Pollack JR, Neilands JB (1970) Enterobactin, an iron transport compound from Salmonella typhimurium . Biochem Biophys Res Commun 38: 989–992. [DOI] [PubMed] [Google Scholar]
- 11. Loomis LD, Raymond KN (1991) Solution equilibria of enterobactin and metal-enterobactin complexes. Inorg Chem 30: 906–911. [Google Scholar]
- 12. Wilson MK, Abergel RJ, Raymond KN, Arceneaux JEL, Byers BR (2006) Siderophores of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis . Biochem Biophys Res Commun 348: 320–325. [DOI] [PubMed] [Google Scholar]
- 13. Dertz EA, Xu J, Stintzi A, Raymond KN (2006) Bacillibactin-mediated iron transport in Bacillus subtilis . J Am Chem Soc 128: 22–23. [DOI] [PubMed] [Google Scholar]
- 14. Bister B, Bischoff D, Nicholson GJ, Valdebenito M, Schneider K, et al. (2004) The structure of salmochelins: C-glucosylated enterobactins of Salmonella enterica . BioMetals 17: 471–481. [DOI] [PubMed] [Google Scholar]
- 15. Sandy M, Butler A (2011) Chrysobactin siderophores produced by Dickeya chrysanthemi EC16. J Nat Prod 74: 1207–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matsuo Y, Kanoh K, Jang J-H, Adachi K, Matsuda S, et al. (2011) Streptobactin, a tricatechol-type siderophore from marine-derived Streptomyces sp. YM5-799. J Nat Prod 74: 2371–2376. [DOI] [PubMed] [Google Scholar]
- 17. Persmark M, Expert D, Neilands JB (1989) Isolation, characterization, and synthesis of chrysobactin, a compound with siderophore activity from Erwinia chrysanthemi . J Biol Chem 264: 3187–3193. [PubMed] [Google Scholar]
- 18. Sandy M, Han A, Blunt J, Munro M, Haygood M, et al. (2010) Vanchrobactin and anguibactin siderophores produced by Vibrio sp. DS40M4. J Nat Prod 73: 1038–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dias GM, Thompson CC, Fishman B, Naka H, Haygood MG, et al. (2012) Genome sequence of the marine bacterium Vibrio campbellii DS40M4, isolated from open ocean water. J Bacteriol 194: 904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Soengas RG, Anta C, Espada A, Paz V, Ares IR, et al. (2006) Structural characterization of vanchrobactin, a new catechol siderophore produced by the fish pathogen Vibrio anguillarum serotype O2. Tetrahedron Letters 47: 7113–7116. [Google Scholar]
- 21. Distel DL, Morrill W, MacLaren-Toussaint N, Franks D, Waterbury J (2002) Teredinibacter turnerae gen. nov., sp. nov., a dinitrogen-fixing, cellulolytic, endosymbiotic gamma-proteobacterium isolated from the gills of wood-boring molluscs (Bivalvia: Teredinidae). Int J Syst Evol Microbiol 52: 2261–2269. [DOI] [PubMed] [Google Scholar]
- 22. Yang JC, Madupu R, Durkin AS, Ekborg NA, Pedamallu CS, et al. (2009) The complete genome of Teredinibacter turnerae T7901: an intracellular endosymbiont of marine wood-boring bivalves (shipworms). PLoS ONE 4: e6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moya A, Peretó J, Gil R, Latorre A (2008) Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat Rev Genet 9: 218–229. [DOI] [PubMed] [Google Scholar]
- 24. Elshahawi SI, Trindade-Silva AE, Hanora A, Han AW, Flores MS, et al. (2013) Boronated tartrolon antibiotic produced by symbiotic cellulose-degrading bacteria in shipworm gills. Proc Natl Acad Sci USA 110: E295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Waterbury JB, Calloway CB, Turner RD (1983) A cellulolytic nitrogen-fixing bacterium cultured from the gland of deshayes in shipworms (bivalvia: teredinidae). Science 221: 1401–1403. [DOI] [PubMed] [Google Scholar]
- 26. Kester DR, Duedall IW, Connors DN, Pytkowicz RM (1967) Preparation of artificial seawater. Limnol Oceanogr 12: 176–179. [Google Scholar]
- 27. Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 111: 1–61. [Google Scholar]
- 28. Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160: 47–56. [DOI] [PubMed] [Google Scholar]
- 29. Marfey P, Ottesen M (1984) Determination of D-amino acids. I. Hydrolysis of DNP-L-amino acid methyl esters with carboxypeptidase-Y. Carlsberg Res Commun 49: 585–590. [Google Scholar]
- 30. Ansari MZ, Yadav G, Gokhale RS, Mohanty D (2004) NRPS-PKS: a knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res 32: W405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bachmann BO, Ravel J (2009) Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Meth Enzymol 458: 181–217. [DOI] [PubMed] [Google Scholar]
- 32. Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14: 755–763. [DOI] [PubMed] [Google Scholar]
- 33. Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, et al. (2012) The Pfam protein families database. Nucleic Acids Res 40: D290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rausch C, Hoof I, Weber T, Wohlleben W, Huson DH (2007) Phylogenetic analysis of condensation domains in NRPS sheds light on their functional evolution. BMC Evol Biol 7: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Röttig M, Medema MH, Blin K, Weber T, Rausch C, et al. (2011) NRPSpredictor2–a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res 39: W362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, et al. (2011) Geneious v5.4, Available from http://www.geneious.com/.
- 38. Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- 39.Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop. New Orleans, LA. 1–8.
- 40. Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8: 275–282. [DOI] [PubMed] [Google Scholar]
- 41. Walsh CT, Liu J, Rusnak F, Sakaitani M (1990) Molecular studies on enzymes in chorismate metabolism and the enterobactin biosynthetic pathway. Chem Rev 90: 1105–1129. [Google Scholar]
- 42. Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, et al. (1996) A new enzyme superfamily – the phosphopantetheinyl transferases. Chem Biol 3: 923–936. [DOI] [PubMed] [Google Scholar]
- 43. Quadri LEN, Sello J, Keating TA, Weinreb PH, Walsh CT (1998) Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem Biol 5: 631–645. [DOI] [PubMed] [Google Scholar]
- 44. Furrer JL, Sanders DN, Hook-Barnard IG, McIntosh MA (2002) Export of the siderophore enterobactin in Escherichia coli: involvement of a 43 kDa membrane exporter. Mol Microbiol 44: 1225–1234. [DOI] [PubMed] [Google Scholar]
- 45. Brickman TJ, McIntosh MA (1992) Overexpression and purification of ferric enterobactin esterase from Escherichia coli. Demonstration of enzymatic hydrolysis of enterobactin and its iron complex. J Biol Chem 267: 12350–12355. [PubMed] [Google Scholar]
- 46. de Lorenzo V, Wee S, Herrero M, Neilands JB (1987) Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol 169: 2624–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Baichoo N, Helmann JD (2002) Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J Bacteriol 184: 5826–5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Conti E, Stachelhaus T, Marahiel MA, Brick P (1997) Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 16: 4174–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stachelhaus T, Mootz HD, Marahiel MA (1999) The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol 6: 493–505. [DOI] [PubMed] [Google Scholar]
- 50. Challis GL, Ravel J, Townsend CA (2000) Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem Biol 7: 211–224. [DOI] [PubMed] [Google Scholar]
- 51. Distel DL, DeLong EF, Waterbury JB (1991) Phylogenetic characterization and in situ localization of the bacterial symbiont of shipworms (Teredinidae: Bivalvia) by using 16S rRNA sequence analysis and oligodeoxynucleotide probe hybridization. Appl Environ Microbiol 57: 2376–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Distel DL, Beaudoin DJ, Morrill W (2002) Coexistence of multiple proteobacterial endosymbionts in the gills of the wood-boring bivalve Lyrodus pedicellatus (Bivalvia: Teredinidae). Appl Environ Microbiol 68: 6292–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chatterjee C, Paul M, Xie L, van der Donk WA (2005) Biosynthesis and mode of action of lantibiotics. Chem Rev 105: 633–684. [DOI] [PubMed] [Google Scholar]
- 54. Bagley MC, Dale JW, Merritt EA, Xiong X (2005) Thiopeptide Antibiotics. Chem Rev 105: 685–714. [DOI] [PubMed] [Google Scholar]
- 55. Nishizawa T, Asayama M, Fujii K, Harada K, Shirai M (1999) Genetic analysis of the peptide synthetase genes for a cyclic heptapeptide microcystin in Microcystis spp. J Biochem 126: 520–529. [DOI] [PubMed] [Google Scholar]
- 56. Nishizawa T, Ueda A, Asayama M, Fujii K, Harada K, et al. (2000) Polyketide synthase gene coupled to the peptide synthetase module involved in the biosynthesis of the cyclic heptapeptide microcystin. J Biochem 127: 779–789. [DOI] [PubMed] [Google Scholar]
- 57. Tillett D, Dittmann E, Erhard M, von Döhren H, Börner T, et al. (2000) Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide–polyketide synthetase system. Chem Biol 7: 753–764. [DOI] [PubMed] [Google Scholar]
- 58. Homann VV, Sandy M, Tincu JA, Templeton AS, Tebo BM, et al. (2009) Loihichelins A-F, a suite of amphiphilic siderophores produced by the marine bacterium Halomonas LOB-5. J Nat Prod 72: 884–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hoffmann K, Schneider-Scherzer E, Kleinkauf H, Zocher R (1994) Purification and characterization of eucaryotic alanine racemase acting as key enzyme in cyclosporin biosynthesis. J Biol Chem 269: 12710–12714. [PubMed] [Google Scholar]
- 60. Bachhawat AK, Ghosh S (1987) Iron transport in Azospirillum brasilense: role of the siderophore spirilobactin. Microbiology 133: 1759–1765. [Google Scholar]
- 61. Scarrow RC, Ecker DJ, Ng C, Liu S, Raymond KN (1991) Iron(III) coordination chemistry of linear dihydroxyserine compounds derived from enterobactin. Inorg Chem 30: 900–906. [Google Scholar]
- 62. Betcher MA, Fung JM, Han AW, O'Connor R, Seronay R, et al. (2012) Microbial distribution and abundance in the digestive system of five shipworm species (Bivalvia: Teredinidae). PLoS ONE 7: e45309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kloepper JW, Leong J, Teintze M, Schroth MN (1980) Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 286: 885–886. [Google Scholar]
- 64. Rock JL, Nelson DR (2006) Identification and characterization of a hemolysin gene cluster in Vibrio anguillarum . Infect Immun 74: 2777–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Simon R, Priefer U, Pühler A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotech 1: 784–791. [Google Scholar]
- 66. Milton DL, O'Toole R, Horstedt P, Wolf-Watz H (1996) Flagellin A is essential for the virulence of Vibrio anguillarum . J Bacteriol 178: 1310–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maximum likelihood tree of C domains, showing the grouping of different C domain subtypes. The C domains of TnbF are shown in red. TnbF's M2 C domain groups with the DCL functional group, while the M1 C domain groups with the Starter functional group. Each C domain is labeled with the organism name, followed by accession number, followed by the module number from which the C domain is referring. C domains from glycopeptide antibiotic NRPS's are shown in blue. Bootstrap values are based on 100 replicates and are only shown for the basal branches of groups.
(TIF)
HPLC trace of HP20 extract from T. turnerae T7901 culture supernatant, recorded at 215 nm.
(TIF)
HPLC trace of derivatized hydrolysate of turnerbactin and chiral amino acid standards, recorded at 340 nm. (A) hydrolysate of turnerbactin, (B) L-ornithine, (C) DL-ornithine, (D) L-serine, (1) D-ornithine, (2) L-ornithine, (3) L-serine, (4) Marfey's reagent (FDAA).
(TIF)
1 1H NMR spectrum (800 MHz) in CD3OD.
(TIF)
1 13C NMR spectrum (800 MHz) in CD3OD.
(TIF)
1 1H-13C HSQC spectrum (800 MHz) in CD3OD.
(TIF)
1 1H-13C HMBC spectrum (800 MHz) in CD3OD.
(TIF)
1 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
1 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
1 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
1 1H-1H TOCSY spectrum (800 MHz) in CD3OD.
(TIF)
1 1H-1H TOCSY spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
1 1H-1H TOCSY spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
1 1H-1H TOCSY spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
2 1H NMR spectrum (800 MHz) in CD3OD.
(TIF)
2 1H-13C HSQC spectrum (800 MHz) in CD3OD.
(TIF)
2 1H-13C HMBC spectrum (800 MHz) in CD3OD.
(TIF)
2 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
2 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
2 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
3 1H NMR spectrum (800 MHz) in CD3OD.
(TIF)
3 1H-13C HSQC spectrum (800 MHz) in CD3OD.
(TIF)
3 1H-13C HMBC spectrum (800 MHz) in CD3OD.
(TIF)
3 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
3 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
3 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
4 1H NMR spectrum (800 MHz) in CD3OD.
(TIF)
4 13C NMR spectrum (800 MHz) in CD3OD.
(TIF)
4 1H-13C HMBC spectrum (800 MHz) in CD3OD.
(TIF)
4 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
4 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
4 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
4 1H-1H TOCSY spectrum (800 MHz) in CD3OD.
(TIF)
4 1H-1H TOCSY spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
4 1H-1H TOCSY spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
4 1H-1H TOCSY spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
5 1H NMR spectrum (800 MHz) in CD3OD.
(TIF)
5 13C NMR spectrum (800 MHz) in CD3OD.
(TIF)
5 1H-13C HMBC spectrum (800 MHz) in CD3OD.
(TIF)
5 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
5 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
5 1H-13C HMBC spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
5 1H-1H TOCSY spectrum (800 MHz) in CD3OD.
(TIF)
5 1H-1H TOCSY spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
5 1H-1H TOCSY spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
5 1H-1H TOCSY spectrum (800 MHz) in CD3OD, expanded region.
(TIF)
NMR data for (DHB-Orn-Ser)2 (2), Dehydrated (DHB-Orn-Ser)2 (4), and Dehydrated (DHB-Orn-Ser)3 (5) (800 MHz) in CD3OD.
(PDF)