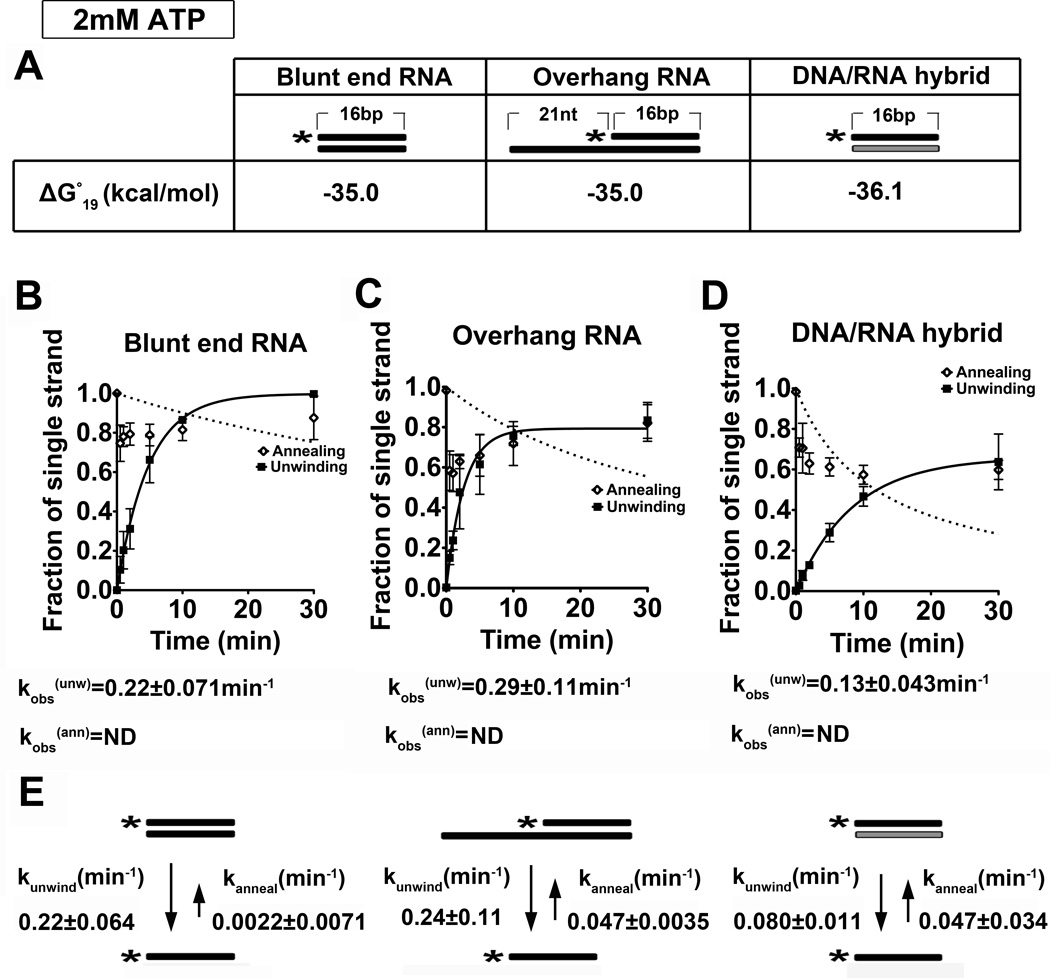
Figure 1. Dbp2 displays ATP-dependent duplex unwinding on multiple RNA substrates at 2 mM ATP:Mg2+.
(A) Schematic representation and thermodynamic stability of RNA duplex substrates. All RNA substrates were designed with similar stability, which was calculated using the Nearest-Neighbor Database and converting to ΔG°19 using ΔG°= ΔH°-T ΔS° 79; 80. Black or gray coloring denotes RNA or DNA strands, respectively, whereas asterisks mark the position of the 32P-radiolabeled 5’ end. (B) Graphical representations of unwinding and annealing assays using radiolabeled 16-bp blunt end RNA duplexes, (C) 21-nt overhang that is 3’ to the 16-bp RNA duplexes and (D) 16-bp blunt end DNA/RNA hybrids. Reactions were performed at 19 °C with 2 mM ATP:Mg2+, 0.1 nM radiolabeled duplex, and 600 nM recombinant, purified Dbp2. The fraction of the single stranded substrate at each time point is plotted as the average of three independent reactions with standard deviations from the mean. The data was fitted to the integrated form of a homogenous first order rate law to determine the kobs(unw). kobs(ann) was determined using the integrated rate law for the bimolecular annealing reaction as previously described 26. N.D. = not determined. Representative non-denaturing gels are shown in Fig S2. (E) Kinetic parameters for Dbp2 unwinding and annealing at 2 mM ATP. The rate constants for Dbp2 unwinding and annealing were calculated as previously described26. This reveals that Dbp2 preferentially unwinds RNA-RNA duplexes irrespective of the presence of a single-stranded overhang region. Dbp2 exhibits RNA-DNA duplex unwinding but with a lower activity than RNA-RNA substrates of similar overall stability.