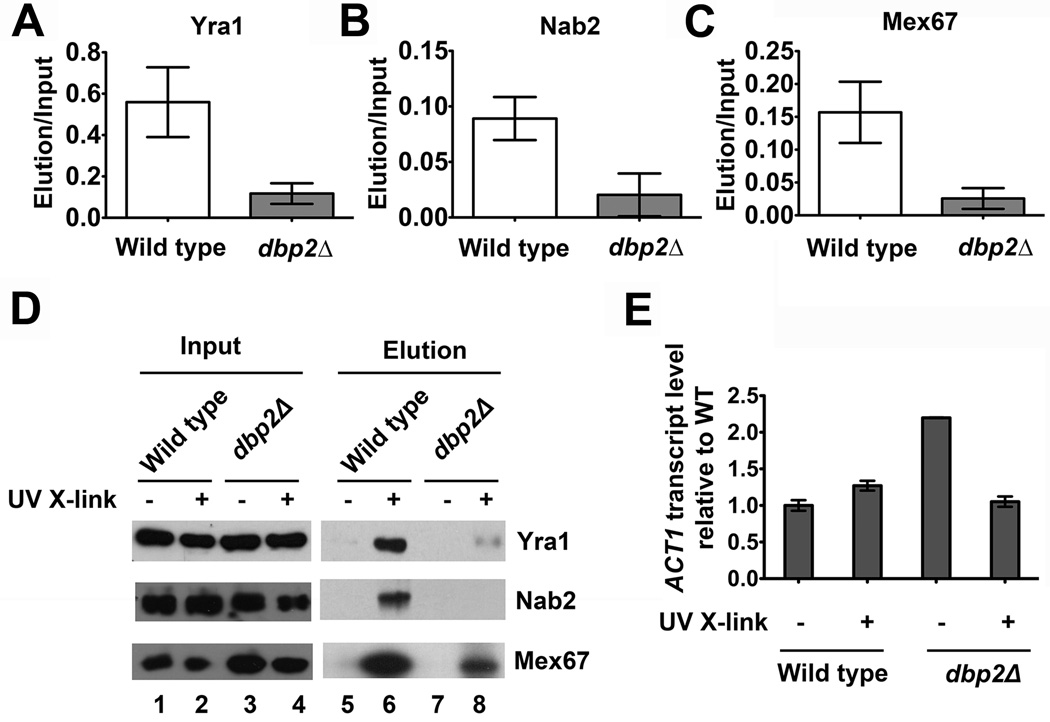
Figure 4. Loss of DBP2 results in reduced association of Yra1, Nab2 and Mex67 with poly(A)+ RNA.
In vivo UV crosslinking reveals reduced association of (A) Yra1, (B) Nab2 and (C) Mex67 with poly(A)+ RNA in dbp2Δ cells. Wild type and dbp2Δ cells were subjected to UV crosslinking followed by poly(A)+ RNA isolation as previously described 55. The eluted fraction of wild type and dbp2Δ cells were normalized to equal RNA concentration using equivalent A260nm absorbance units. Proteins from the eluted fractions were detected by Western blotting. The relative quantity of poly(A)+ RNA-bound proteins was determined following quantification of the resulting isolated proteins from three independent biological replicates and is reported as the amount of isolated protein relative to total (input). (D) Representative western blot of in vivo UV crosslinking. The total protein abundance (input) is shown along with the amount of isolated proteins with and without UV crosslinking. The latter serves as a background control to show that proteins isolated following UV crosslinking are not due to non-specific interactions. (E) Reverse-transcriptase, quantitative PCR (RT-qPCR) shows efficient isolation of ACT1 mRNA from both wild type and dbp2Δ cells following oligo-dT selection. Equal fractions of eluted RNA were reverse transcribed and subjected to qPCR with ACT1-specific primers as previously described 18. Transcript levels were normalized by setting the wild type elution without UV crosslinking to 1 and are a result of three technical replicates from one biological sample per strain.