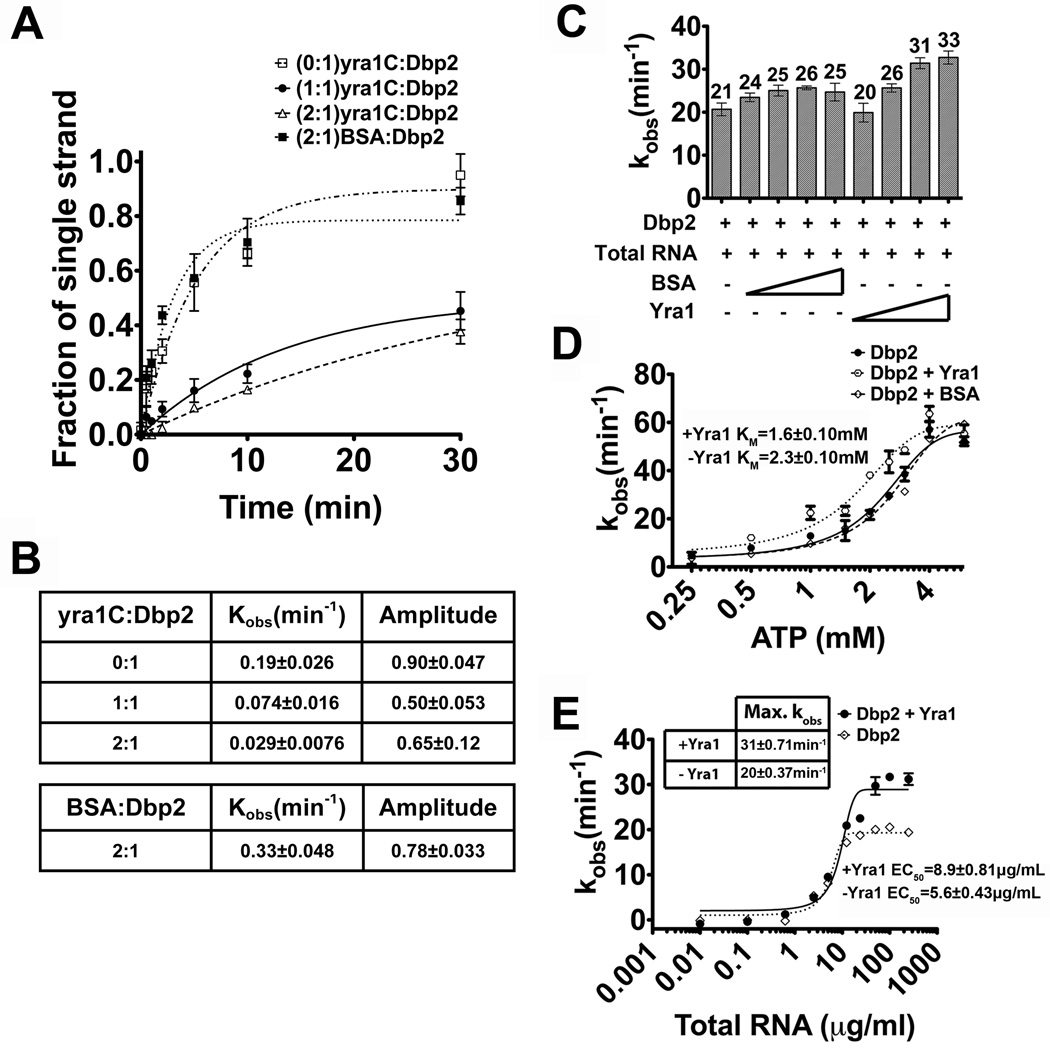
Figure 6. Yra1 modulates the enzymatic activity of Dbp2.
(A) Graphical representation of Dbp2 duplex unwinding with yra1C. Unwinding assays were conducted with the blunt end RNA duplex and either Dbp2 alone (600 nM) or with yra1C (600 nM, 1200 nM) or BSA (1200 nM). Representative non-denaturing gels are shown in Fig S4 and demonstrate that yra1C and BSA do not have intrinsic annealing activity. (B) The kobs(unw) and the amplitude of the unwinding reaction. The kobs(unw) and the amplitude are determined using the integrated rate law for a homogeneous first-order reaction as previously described26(C) Full length Yra1 moderately enhances ATP hydrolysis activity of Dbp2. In vitro ATPase assays were conducted with 200 nM of recombinant, purified Dbp2 and 250 µg/ml of total yeast RNA using a PK/lactate dehydrogenase enzyme-coupled absorbance based detection method as previously described 18. Recombinant, purified Yra1 was included where indicated at final concentrations from 100–600 nM. Equal concentrations of BSA were also tested to account for non-specific interactions. The ATPase activity of Dbp2 alone is similar to previous publications and has already been characterized 18(D) Yra1 moderately enhances the ATP binding affinity of Dbp2. In vitro ATPase assays were conducted as above with constant amounts of Dbp2, Yra1, total RNA (10 µg/mL), increasing amounts of ATP and constant MgCl2 (2 mM). Assays were also conducted with BSA in place of Yra1 to account for non-specific effects. The KM is the indicative of the ATP binding affinity of Dbp2. (E) Yra1 slightly increases the amount of RNA necessary for activation of ATP hydrolysis. In vitro ATPase assays were conducted as above with 200 nM Dbp2, 400 nM Yra1 and increasing amounts of total yeast RNA. The amount of RNA necessary for 50% stimulation of maximum ATPase activity (EC50) is reflective of the RNA binding affinity of Dbp2.