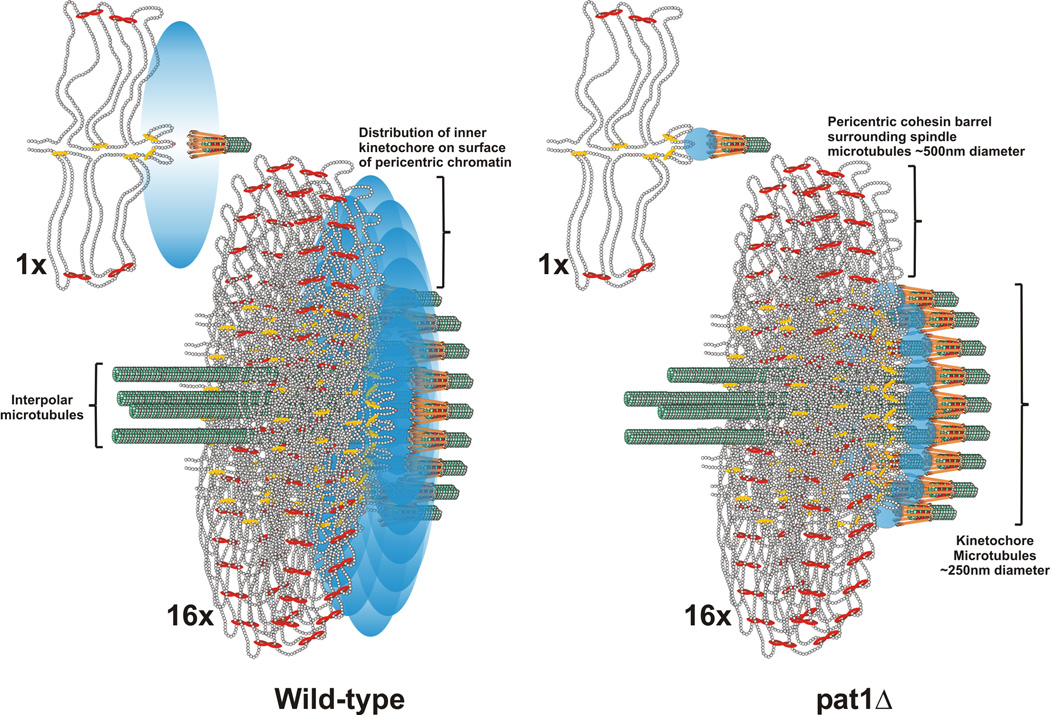
Figure 4. The inner kinetochore is organized into a disc at the surface of the pericentric chromatin in mitosis.
The pericentromere DNA is organized as a network of loops with the centromere DNA at the apex (DNA strands shown as strings of nucleosomes in gray) [11, 14, 23]. The centromere is attached to microtubule plus-ends via the kinetochore (orange barbells surrounding the microtubule in green). Cohesin (red) and condensin (yellow) are enriched in the pericentromere and surround the central spindle [14, 23]. A single pericentric region (one sister) is shown (1X). In metaphase centromeres from the 16 chromosomes are clustered around 16 kinetochore microtubules that emanate from the spindle pole body (16X, below). 4 interpolar microtubules extend to overlap with 4 interpolar from the opposite spindle pole body (not shown). Only the section of the pericentric barrel at the interface of the kinetochore microtubules is shown for clarity. In wild-type, a Cse4 molecule (pink ball) resides at the centromere DNA (apex of the loop)(see 1X, wild-type). Accessory molecules (Pat1 dependent) are in the vicinity, represented as a probability bubble (graded blue oval) at the microtubule plus-end. The emergent spatial arrangement upon clustering of 16 kinetochores (16X) predicts the anisotropy and experimentally generated statistical maps. The width of the density map for a single molecule Cse4 at each CEN and 4 molecules with a probability bubble 550nm around the spindle is 190 ± 141nm (data not shown). This distribution is commensurate with the intensity measurements of deconvolved Cse4 images (Fig. 3, inset). Wild-type density maps of Cse4 have a width of 181 ± 155nm (Fig. 3). In pat1Δ mutants, the accessory molecules are absent, and the remaining Cse4 resides at the kinetochore microtubule plus-end (blue circles, pat1Δ). The predicted geometry in the pat1Δ matches the isotropy and experimentally generated statistical maps (Fig. 2). This arrangement suggests that Cse4 and other inner kinetochore components constitute an inner plate at the chromatin, much like that observed in the trilaminar structure at the chromosome surface of a mammalian kinetochore.